Abstract
Though G-proteins have been implicated in the primary step of taste signal transduction, no direct demonstration has been done in insects. We show here that a G-protein gamma subunit, Gγ1, is required for the signal transduction of sugar taste reception in Drosophila. The Gγ1 gene is expressed mainly in one of the gustatory receptor neurons. Behavioral responses of the flies to sucrose were reduced by the targeted suppression of neural functions of Gγ1-expressing cells using neural modulator genes such as the modified Shaker K+ channel (EKO), the tetanus toxin light chain or the shibire (shits1) gene. RNA interference targeting to the Gγ1 gene reduced the amount of Gγ1 mRNA and suppressed electrophysiological response of the sugar receptor neuron. We also demonstrated that responses to sugars were lowered in Gγ1 null mutant, Gγ1N159. These results are consistent with the hypothesis that Gγ1 participates in the signal transduction of sugar taste reception.
Keywords: Drosophila, G-protein, signal transduction, taste
Introduction
Gustatory sense enables animals to distinguish among nutritious and toxic soluble substances. Although gustatory sense plays a key role in determining the taste of food, the signaling mechanisms governing this important sensory system are not fully understood. As a model system, Drosophila offers several advantages for exploring the mechanism of gustatory sense perception at different levels of an organism (Ishimoto and Tanimura, 2004). In Drosophila, as in other insects, taste substances are detected by bipolar gustatory receptor neurons (GRNs). A typical chemosensilla houses four kinds of GRNs. Each GRN works as a specialist characterized by its responsiveness to taste substances. Sugar cells (S cells) respond to mono-, di- and trisaccharides. W and L1 cells respond to water and low concentration of salt, respectively. L2 cells respond to deterrent stimulants such as high concentrations of salt and bitter compounds (Meunier et al, 2003).
Previous studies have indicated that sugar taste information is initially received by G-protein-coupled receptors, both in mammals and insects (Hoon et al, 1999; Clyne et al, 2000; Dunipace et al, 2001; Nelson et al, 2001, 2002; Scott et al, 2001; Li et al, 2002). A typical model of G-protein signaling involves G-protein-coupled receptors coupled to a membrane-associated heterotrimer composed of a GTP-hydrolyzing Gα subunit and a Gβγ dimeric partner. G-protein subunits that mediate intracellular taste signaling pathways have been identified only in mammals (Lindemann, 2001). Several mammalian molecules, including G-protein subunits, a phospholipase C (PLC), a phosphodiesterase (PDE), an inositol 1,4,5-triphosphate (Ins(1,4,5)P3) receptor and a transient receptor potential-like (TRPL) channel, have been linked to the taste transduction pathway. One of the G-protein α subunits, α-gustducin (Mclaughlin et al, 1992), is known to be involved in the mammalian response to sweet and bitter compounds (Wong et al, 1996). Gγ13, a G-protein γ subunit, is involved in a signal transduction pathway for a bitter compound, denatonium (Huang et al, 1999). Phospholipase C-β2 (PLCβ2) is essential for sweet and bitter signal transduction pathways (Zhang et al, 2003). A TRPL channel, TRPM5, is involved in both sweet and bitter taste signaling pathways (Zhang et al, 2003). On the other hand, taste signal transduction pathways in insects remain to be determined. In the visual system, the signal transduction pathway is divergent between vertebrates and invertebrates (Hardie and Raghu, 2001). In the vertebrate phototransduction system, a G-protein α subunit, transducin, activates a PDE, resulting in hydrolysis of guanosine 3′,5′-cyclic monophosphate and closure of transduction channels. In Drosophila, rhodopsin activates a distinct G-protein isoform, dGqα (Talluri et al, 1995), which activates a PLC isoform encoded by the norpA gene. Thus, Drosophila phototransduction employs an Ins(1,4,5)P3 pathway instead of a cGMP pathway for the signal transduction. The Ins(1,4,5)P3 pathway likely plays a role in the signal transduction cascade eliciting sweet taste in the fleshfly, Boettcherisca peregrine (Koganezawa and Shimada, 2002). To understand the molecular mechanisms underlying taste reception, additional information on the signal transduction molecules is needed both in vertebrates and invertebrates. In this study, we determined that a G-protein γ subunit, Gγ1, is expressed in GRNs and demonstrated that Gγ1 is involved in responses to sugars using the behavioral and electrophysiological analysis combined with molecular genetic techniques in Drosophila.
Results and discussion
G-protein subunits are expressed in gustatory sensory organs
In the Drosophila genome, 16 genes are predicted to encode G-protein subunits. These include sequences for 11 α subunits, three β subunits and two γ subunits (FlyBase). We determined G-protein subunits expressed in a gustatory organ using RT–PCR. Gα73B, Gβ13F, Gβ5 and Gγ1 were detected by this analysis (Supplementary Figure S1). The role of these G-protein subunits in gustatory signal transduction has not been shown previously. In this study, we have characterized one of these four G-proteins, Gγ1. To study the role of G-proteins involved in signal transduction of taste, we chose the G-protein γ subunit, since Drosophila G-protein γ subunit includes the fewest variety of subtypes, only two: Gγ1 and Gγ30A, among three G-protein subunits and only one subtype, Gγ1, is expressed in the gustatory organ labellum (Figure 1A). Gγ1 (CG8261) is located on the second chromosome and is cytologically mapped to 44F3–5. There are five alternative transcriptional forms (RA–RE) of the Gγ1 gene. To identify which transcripts are expressed in the labellum, we carried out RT–PCR and 3′ RACE using specific primers for these transcripts (Supplementary Figure S2). We found that the RC, RD and RE forms are present in labellum. We searched Gal4 enhancer-trap strains to study the role of Gγ1 in the gustatory system (Brand and Perrimon, 1993). We found NP1535 strain in which a P{Gal4} element is inserted 73 bp upstream of the transcriptional start site of Gγ1 (Figure 1B) (Ray and Ganguly, 1992, 1994). The UAS-green fluorescent protein (GFP) and NP1535 strains were crossed and their progeny enabled us to visualize Gγ1 expression with GFP detection. GFP signals were observed in both the labellum and the tarsi (Figure 1C). Gustatory chemosensilla on the labellum are classified into three types: l-, s- and i-type, based on their shape and location (Ishimoto and Tanimura, 2004). Four GRNs are housed in s- and l-type chemosensilla as a cluster, while the i-type chemosensilla house two GRNs (Hiroi et al, 2004). We observed GFP signals in all GRNs from each cluster, though only one GRN exhibited a higher GFP signal at the high magnification (Figure 1C inset), suggesting that Gγ1 mainly functions in one of the four GRN types.
Figure 1.
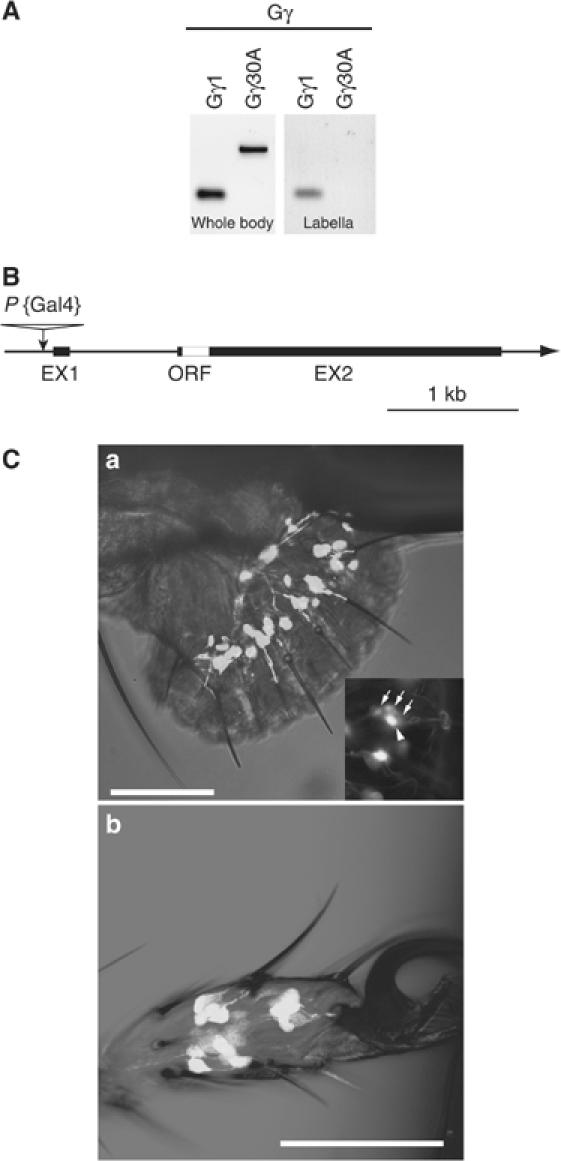
G-protein gamma subunit 1 is expressed in gustatory organ. (A) RT–PCR analysis shows an mRNA expression profile of two G-protein gamma subunits in the labellum and whole body. (B) A schematic diagram showing the gene structure of the Gγ1 gene and the insertion site of P{Gal4} in the enhancer trap strain, NP1535. (C) GAL4 expression patterns in NP1535 visualized by GFP. GFP expression is in the sensory neurons of labellum (a) and tarsi (b). GFP signals were observed at the base of all gustatory sensilla in the labellum and tarsi. In a cluster of four GRNs, one GRN shows higher intensity of GFP signal. Arrows indicate GRNs showing lower intensity of GFP signals in a cluster of GRNs. An arrowhead indicates a cell with a higher intensity of GFP signal. Scale bars indicate 50 μm.
Neural suppression directed by NP1535 Gal4 driver reduces behavioral response to sucrose
We tested the possible role of Gγ1 in gustatory signal transduction by measuring the proboscis extension reflex (PER) response to sucrose, since G-protein-coupled receptors are known to mediate the sweet signal transduction pathway both in vertebrates and invertebrates (Lindemann, 2001). As a control strain, we used ΔP NP1535#1–3, in which a P{Gal4} element was precisely removed by introducing a genomic transposase source. ΔP NP1535 flies showed an 83–91% PER response rate (Figure 2A), whereas the NP1535 homozygote flies exhibited significantly lowered PER rates (P<0.001). The NP1535/ΔP NP1535#1 heterozygote flies, like the ΔP NP1535 flies, showed normal PER. The P{Gal4} insertion induced a recessive phenotype in the behavioral response to sucrose. To compare the expression level of Gγ1 mRNA between NP1535 and control strains, we performed quantitative PCR (QPCR) (Figure 4B). The results indicated that Gγ1 expression level is actually reduced in NP1535 flies in which the amount of Gγ1 mRNA is 9–14% of control strains (Figure 4B). To characterize Gγ1-expressing GRNs, we examined the PER response of flies expressing mutant type of generically modified Shaker K+ channels (Osterwalder et al, 2001; White et al, 2001), EKO, driven by the Gal4 reporter of NP1535. In the NP1535/+; UAS-EKO/+ flies, EKO inhibits neural activity of the Gal4-expressing cells (Figure 2A). The NP1535/+; UAS-EKO/+ flies exhibited a significantly lowered PER rate of 36% (P<0.001), which is similar to the response of the NP1535 homozygote flies. Tetanus toxin (TNT) (Sweeney et al, 1995) inhibits docking of the synaptic vesicles to the membrane, thus blocking neurotransmitter release. The PER response of flies expressing TNT was also measured (Figure 2A). In the NP1535/UAS-TNT flies, the PER responses were severely reduced, even in comparison to that of the NP1535/+; UAS-EKO/+ flies. These results suggest that the Gal4-expressing neurons in the NP1535 strain participate in the behavioral response to sucrose. However, there remains a possibility that the action of EKO or TNT might impair developmental processes in the nervous systems regulating the observed PER response.
Figure 2.
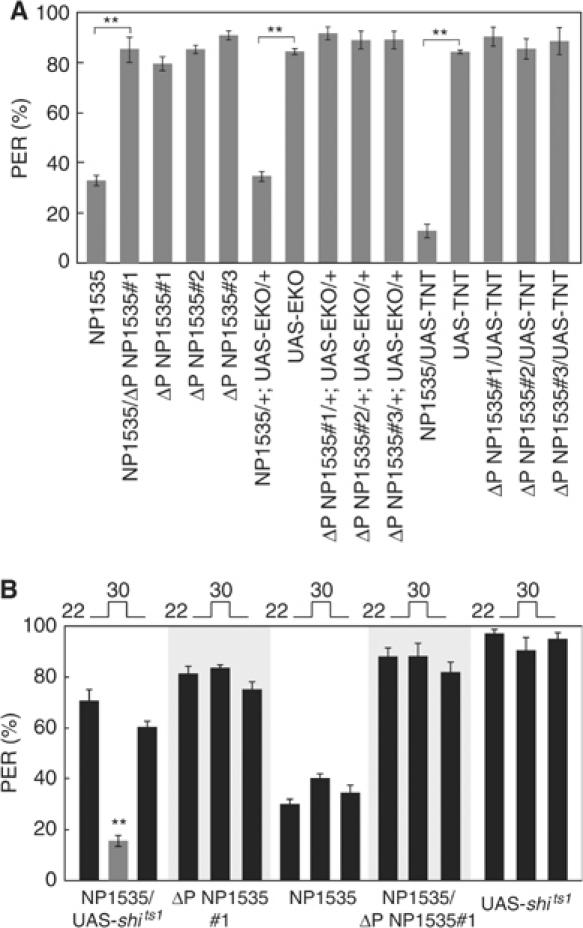
PER is suppressed by targeted expression of EKO or TNT. (A) PER responses of flies expressing either UAS-EKO or UAS-TNT induced by enhancer-trap line (NP1535) and control flies (NP1535, UAS-EKO, UAS-TNT, ΔP NP1535#1–3, NP1535/ΔP NP1535#1) to 100 mM sucrose. **P<0.001. (B) PER responses of NP1535/UAS-shits1 flies and control flies (NP1535, UAS-shits1, ΔP NP1535#1, NP1535/ΔP NP1535#1) to 100 mM sucrose at the permissive temperature (22°C) and restrictive temperature (30°C). Data were obtained from at least 150 flies of each strain. **P<0.001.
Figure 4.
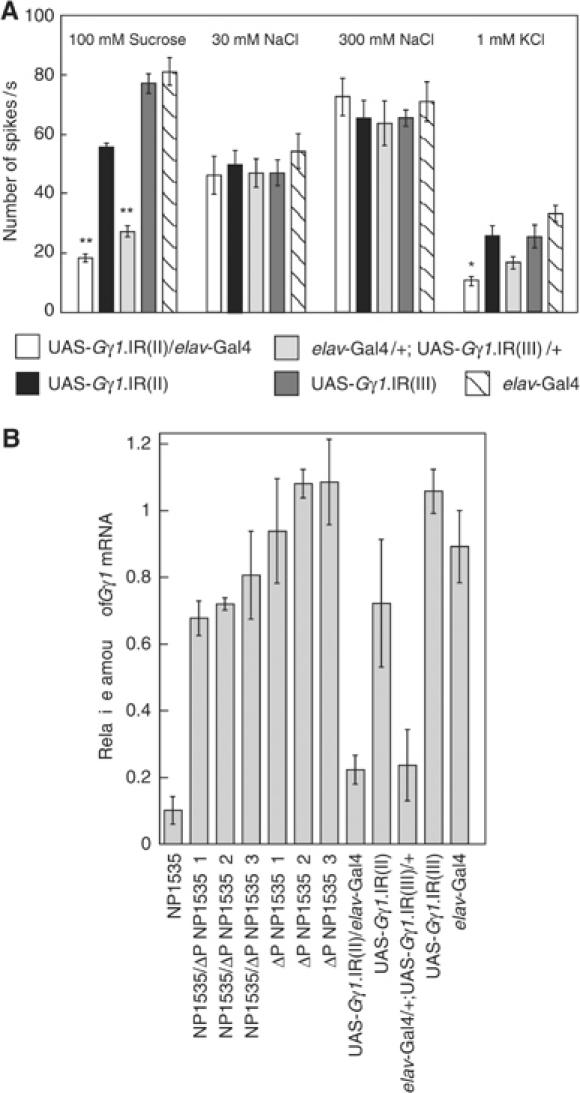
RNAi-mediated Gγ1 silencing impairs nerve responses of GRNs to sugar. (A) Nerve responses were recorded from l-type chemosensilla on the labellum. Stimulating solutions were 100 mM sucrose for S cell, 30 mM NaCl for L1 cell, 300 mM NaCl for L2 cell and 1 mM KCl for W cell. We used UAS-Gγ1.IR flies with either the second or third chromosome linked. We obtained responses from at least 10 flies. Error bars are s.e.m.; **P<0.001; *P<0.01 (B) mRNA expression levels were measured by QPCR. Elongation factor 1 α48D (Ef1α48D:CG8280) was used to normalize the mRNA content among strains. Error bars are s.e.m.; **P<0.001; *P<0.01.
To exclude this possibility, we used a temperature-sensitive allele of the dynamin mutant, shits1, through which synaptic transmissions can be conditionally disrupted (Figure 2B). At a restrictive temperature (30°C), the neurotransmitter release is inhibited in the shits1-expressing neurons. The NP1535/UAS-shits1 flies showed normal PER responses to 100 mM sucrose at the permissive temperature (22°C). When these flies were transferred to 30°C, the PER response was significantly reduced (P<0.001). This reduction was recovered when flies were returned to 22°C. The control flies, NP1535, ΔP NP1535#1, NP1535/ΔP NP1535#1 and UAS-shits1, showed no significant differences of PER responses under both temperatures (P>0.1). These data suggest that the Gal4-expressing neurons in the NP1535 flies are required for the behavioral response to sucrose.
Nerve responses to sugars were reduced in NP1535/UAS-EKO and NP1535 flies
We showed that the reduced behavioral response to sucrose is due to suppression of the Gal4-expressing neurons in NP1535. Yet, we were not able to discern whether the cause of the behavioral defect is in the peripheral nervous system or in the central nervous system.
We then recorded the electrophysiological GRN response to determine whether the Gγ1-expressing GRNs are needed for the sugar reception. We found no significant reduction of nerve responses to water and salt in NP1535 and NP1535/+; UAS-EKO/+ flies (data not shown; P>0.1). In the ΔP NP1535 and UAS-EKO flies, we observed approximately 64 spikes/s from the labellum gustatory sensilla using 100 mM sucrose (Figure 3A), whereas the NP1535 flies showed a decreased firing rate (38 spikes/s). This result is consistent with the behavioral data obtained by the PER test (Figure 2). The NP1535/+; UAS-EKO/+ flies also demonstrated an attenuated nerve response (Figure 3A). These results support the view that Gγ1 is functioning in S cells.
Figure 3.
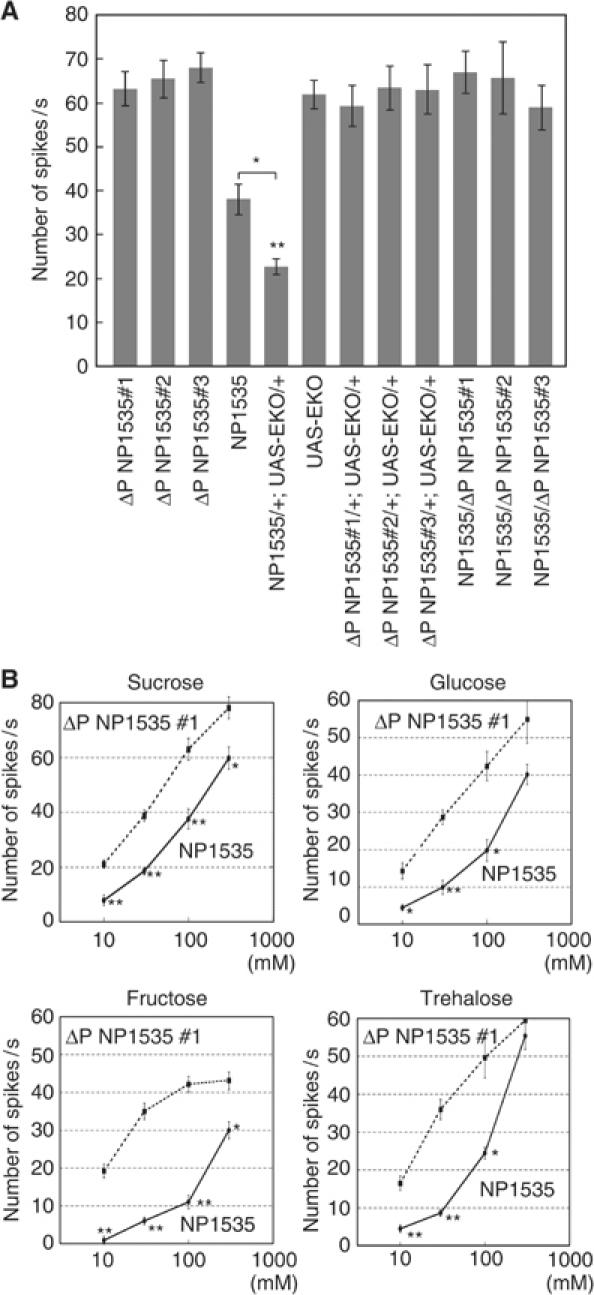
Nerve responses were reduced in NP1535 and NP1535/+; UAS-EKO/+ flies. (A) Nerve responses of NP1535/+; UAS-EKO/+ flies to 100 mM sucrose. Responses were recorded from l-type chemosensilla on the labellum. Significant differences were observed between control flies (ΔP NP1535#1–3, NP1535/ΔP NP1535#1–3, ΔP NP1535#1–3/+;UAS-EKO/+ and UAS-EKO) and NP1535/+; UAS-EKO/+ flies. Nerve responses were obtained from 10 flies. Error bars are s.e.m.; **P<0.001; *P<0.05. (B) Dose–response curves of nerve responses to sugars (sucrose, glucose, fructose and trehalose) of NP1535 and ΔP NP1535#1 flies. Each data point represents at least five recordings made from 12 flies in total. Error bars are s.e.m.; **P<0.001; *P<0.05.
S cells respond to sugars with a glycopyranoside moiety, as well as fructose and trehalose (Rodrigues and Siddiqi, 1981; Tanimura and Shimada, 1981; Tanimura et al, 1982). We examined the neural responses of flies to four kinds of sugars: sucrose, glucose, fructose and trehalose, at a range of concentration from 10 to 300 mM (Figure 3B). In comparison to the control flies, the NP1535 flies showed markedly lower responses to all four kinds of sugars (10–100 mM of sucrose and fructose; P<0.001 at low sugar concentrations, 30 mM of glucose; P<0.001, 10 and 30 mM of trehalose; P<0.001). These results are in agreement with the nerve responses we recorded from the NP1535/+; UAS-EKO/+ flies in the present study. The lower Gγ1 expression levels in the NP1535 flies likely cause the attenuation of sugar responses. We used RNA interference (RNAi) methodologies to verify the lower Gγ1 expression response on another genetic background. Moreover, we examined fly nerve responses to water and salt to ask whether the Gγ1 subunit specifically mediates the gustatory signal transduction utilized for sugar perception.
RNAi of Gγ1 suppressed nerve responses of S cells
We examined the nerve responses of flies expressing double-stranded RNA targeting to Gγ1 mRNA (Gγ1.IR) in all neurons using the elav-Gal4 activator. For these studies, we used two independent UAS-Gγ1.IR strains in which a P{UAS-Gγ1.IR} is on the second (II) or the third (III) chromosome. The UAS-Gγ1.IR(II)/elav-Gal4 flies showed reduced nerve responses to 100 mM sucrose (Figure 4A). We also observed reduced nerve responses to 100 mM sucrose in the elav-Gal4/+; UAS-Gγ1.IR(III)/+ flies. The Gγ1 response values using the RNAi methodology were similar to those we observed in NP1535/+; UAS-EKO/+ and NP1535 flies. We used 30 and 300 mM sodium chloride to examine the nerve responses of L1 and L2 cells, respectively. The L1 and L2 cells showed normal responses to sodium chloride. These results indicate that inhibition of the Gγ1 mRNA reduced the S-cell nerve responses, but not those of the L1 (low salt) or L2 (high salt) cells. We also examined nerve responses to water (1 mM KCl) using UAS-Gγ1.IR(II)/elav-Gal4 flies and elav-Gal4/+; UAS-Gγ1.IR(III)/+ flies. The elav-Gal4/+; UAS-Gγ1.IR(III)/+ and the background strain flies responded to water similarly (P>0.1). However, the water response exhibited by the UAS-Gγ1.IR(II)/elav-Gal4 flies was reduced by comparison. To demonstrate the function of Gγ1 in water reception, water response of Gγ1 null mutant should be tested. The RNAi assay for Gγ1 revealed that Gγ1 mediates signaling at some step for sugars. However, the observed nerve responses to sucrose were not totally inhibited in the Gγ1.IR flies, suggesting a possibility that the RNAi was not complete. We determined the amount of Gγ1 mRNA in each strain using QPCR (Figure 4B). Gγ1.IR flies, UAS-Gγ1.IR(II)/elav-Gal4 and elav-Gal4/+;UAS-Gγ1.IR(III)/+, showed reduced expression of Gγ1, which was 20–29% of UAS-Gγ1.IR(II), UAS-Gγ1.IR(III) and elav-Gal4 strains. QPCR results suggest that Gγ1 mRNA was not completely abolished, but substantially reduced by RNAi.
Cells homozygous for Gγ1N159 show reduced sugar responses
We examined nerve responses of GRNs bearing a nonsense mutation Gγ1N159 (Izumi et al, 2004), to determine whether the sugar response will be completely disappeared. Since Gγ1N159 homozygote mutants are embryonic lethal, we employed a directed mosaic system using a GAL4-responsive yeast site-specific recombinase, called FLP (flippase). In this system, a clonal analysis can be restricted to the tissue of interest (Duffy et al, 1998). We combined the elav-GAL4 driver with the UAS-flp responder for the directed recombination of neural precursors including progenitor cells of GRNs. Directed FLP expression then induces mitotic recombinations and generates cells containing the Gγ1N159 homozygotes. We found that GRNs bearing homozygous Gγ1N159 mutants exhibited normal responses to water, high (300 mM) and low (30 mM) concentrations of salt (Figure 5). These results suggest that Gγ1 participates neither in water nor salt reception mechanisms. The reduction of the W-cell response of the UAS-Gγ1.IR(II)/elav-Gal4 flies was possibly caused by an effect of genetic background.
Figure 5.
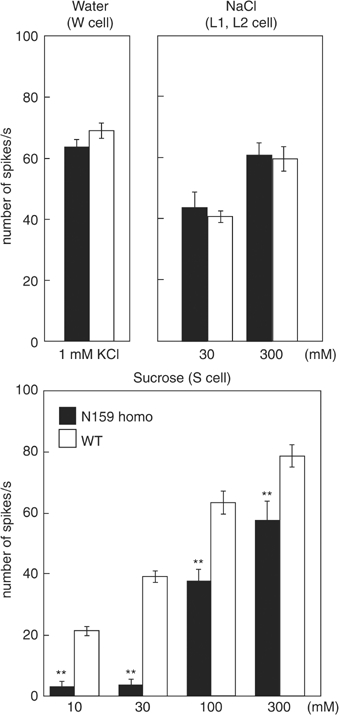
Nerve responses recorded from homozygotes of Gγ1 nonsense mutant cells induced by mitotic recombination. Nerve responses were recorded from normal and somatic recombinant GRNs of l-type chemosensilla. We recorded electrophysiological responses from GRNs whose genotypes were WT or nonsense mutants of the Gγ1 gene (N159 homo). Concentration of sucrose ranged from 10 to 300 mM. In all, 1 mM of KCl was used for stimuli to W (water) cells; 30 and 300 mM of NaCl were used for stimuli to L1 (low salt) and L2 (high salt) cells, respectively. Each recording was obtained from at least 32 chemosensilla of 14 flies. Error bars are s.e.m.; **P<0.001.
GRNs bearing the wild-type (WT) Gγ1 gene responded to sucrose normally (Figure 5). GRNs bearing the homozygous Gγ1N159 mutation showed lower nerve responses to 10–300 mM sucrose. The neural activity in response to the low concentration of sucrose was significantly reduced in the Gγ1N159 homozygote-carrying GRNs (P<0.001), though they still responded to high sucrose concentrations. Therefore, the S cell was not completely suppressed in the Gγ1 null mutant type. There are two G-protein γ subunits in the Drosophila genome. One of the Gγ subunits, Gγ30A, is involved in the phototransduction pathway (Schulz et al, 1999). RT–PCR failed to detect the Gγ30A transcript in the taste organ. Hence, it is unlikely that Gγ30A mediates a sugar reception pathway. A G-protein-independent signal transduction pathway might exist for the sugar signal transduction. In the fleshfly, B. peregrine, Murakami and Kijima (2000) demonstrated that a putative ion channel is directly gated by sucrose using an in situ patch clamp. The null mutant analysis of Gγ1 supports the idea that a G-protein-independent pathway is involved in the sugar reception mechanism.
When we monitored the expression of the Gγ1 gene using the Gal4/UAS system, one of the four GRNs showed the stronger gene expression. Our results proved that only the sugar responses were affected by interfering with the function of the GFP-positive cells. Therefore, it is reasonable to conclude that the cells most strongly labeled by GFP expression are the S cells. However, if Gγ1 is expressed in other GRNs, then there might be an additional role of the gene. One possible function of Gγ1 may be to facilitate the bitter taste signal transduction. The mouse G-protein γ subunit, Gγ13, is colocalized within α-gustducin circumvallate papillae, and is thought to be coupled to mediate sweet and bitter signals (Huang et al, 1999). Since a minority of GRNs are activated by bitter substances (Meunier et al, 2003), there remains a possibility that common G-proteins mediate both sweet and bitter signal transduction pathways in Drosophila. An additional possible function of Gγ1 is a role in developmental processes, since Gγ1 is required for an asymmetrical division of neuroblasts in Drosophila (Izumi et al, 2004). A previous study showed that the expression of Gγ1 is developmentally regulated in a variety of tissues (Ray and Ganguly, 1992). The Gγ1 gene produces at least five transcriptional products. We detected three types of Gγ1 transcripts, RC, RD and RE, in taste organ (Supplementary Figure S2). One or more of these transcripts may function during the development of GRNs.
In this study, we identified Gγ1 expression in the gustatory organs and demonstrated that the Gγ1 subunit is involved in signal transduction pathways of sugar reception. This is the first report that implicates a specific G-protein gamma subunit in this pathway, which is required for sugar taste reception in Drosophila. This finding should lead to a better understanding of the molecular mechanism governing gustatory perception. Potential players include G-protein β subunits associating with Gγ1, downstream signaling molecules and targeted effecters of the Gβ/Gγ1 complex. Our study also alludes to a potential G-protein-independent mechanism of sweet reception. Importantly, this has also been proposed through electrophysiological studies on knockout mice for the G-protein alpha subunit, α-gustducin. α-gustducin knockout mice showed a reduced, but not a completely abolished, response to sweet compounds (He et al, 2004). Thus, a G-protein-independent pathway for sweet taste reception is possibly used in both mammals and insects. Additional studies are needed to identify molecules mediating a G-protein-independent pathway.
Materials and methods
Fly strains
Strains of Drosophila melanogaster were maintained on a standard cornmeal-glucose agar medium at 25°C. A UAS-EKO (the modified Shaker K+ channel) strain was obtained from W Benjamin in the Keshishian lab (Yale University, USA). A UAS-TNT strain was obtained from C O'Kane (Cambridge University, UK). UAS-GFP and elav-Gal4 strains were obtained from the Bloomington Drosophila Stock Center (Indiana, USA). UAS-shibirets1 (Kitamoto, 2001) and Gγ1N159 (Izumi et al, 2004) flies were provided by T Kitamoto (University of Iowa, USA) and F Matsuzaki (Riken, Japan), respectively. The enhancer trap strain, NP1535 (Hayashi et al, 2002), was obtained from the Drosophila Genetic Resource Center in Kyoto Institute of Technology, Japan. For generating flippase-mediated somatic recombinant GRNs carrying the homozygous Gγ1N159 mutation, female flies carrying P elements (P{GawB}elavC155, P{hsflp}1, w*; P{FRT(whs)}G13 P{tubP-Gal80}LL2/CyO, homozygous for hsp70-flp) were crossed to male flies of y1 w*; P{FRT(whs)}G13 P{UAS-mCD8∷GFP.L}LL5, homozygous for {FRT}G13, UAS-mCD8∷GFP (both strains were obtained from the Bloomington Drosophila Stock Center). Male flies carrying both hsp70-flp and UAS-mCD8∷GFP were crossed to female flies carrying Gγ1N159, {FRT}G13 heterozygous with a balancer, CyO. Flies carrying the homozygote form of the Gγ1N159 mosaic GRN clone were generated by inducing heat shock at 30°C three times for 2 h during the mid-pupal stage. Those flies were checked for disappearance of GFP signals under a dissecting microscope equipped with epifluorescence.
RT–PCR analysis
mRNA was prepared from 50 labella (dissected with a razor blade) using a QuickPrep™ Micro mRNA Purification kit (Amersham-Pharmacia Biotech, Uppsala, Sweden). cDNA synthesis and amplification were carried out sequentially using a SuperScript One-Step RT–PCR with Platinum Taq (Invitrogen, CA, USA). Primer sequences were designed to synthesize cDNA sized to 216 bp for Gγ1 (forward: ATGGACGTAATGTCATCATC; reverse, TCCTTAGAGAACGGTGCAGG), and 840 bp for Gγ30A (forward, AGTCGCCCATCCTGCGAAGC; reverse, AGCCTAGATCGAACTCATAC).
Microscopy
The labellum and tarsus of the NP1535/UAS-GFP flies were fixed with phosphate-buffered saline (PBS) containing 4% formaldehyde for 15 min at room temperature. GFP fluorescence was observed with a confocal microscope (Zeiss LSM 510). The excitation wavelength was at 488 nm (argon laser), and the emission wavelength was at 515 nm. Z sections were collected at 1-μm intervals, and were processed to construct projections through an extended depth of focus. Images were processed minimally by using Photoshop (Adobe Systems, California, USA) to adjust the light levels, as well as background contrast and brightness.
Behavioral assays
The PER was examined principally as described (Kimura et al, 1986). Flies aged 3–6 days after eclosion were maintained on fresh medium for 1 day. Flies were starved for 20 h, but were allowed to take water. Before the assay with sugar solutions, the prothoractic tarsus of the fly was touched with a drop of water. If the water droplet induced the PER, the fly was allowed to intake sufficient water. This procedure was repeated between sugar stimulations to prevent water response. We tested PER reactions to sugar solution on 150 flies each, and repeated each experiment three times.
Electrophysiological recordings
Flies 3–6 days old were fed on a fresh medium for 1 day prior to experimentation. All electrophysiological recordings were obtained from labellar chemosensilla using the tip-recording method (Hodgson et al, 1955; Hiroi et al, 2002). Briefly, the proboscis was fixed at the base of labella using lanolin (Wako Pure Chemical Industries, Ltd, Osaka, Japan). The Drosophila Ringer solution, filling the glass capillary tube, grounded the fly subject electrically. Labella chemosensilla were stimulated up to 2 s with a recording electrode with a 20-μm tip diameter. The electrolyte (1 mM KCl) does not elicit spikes from the S, L1, and L2 cells, but elicits spikes from the W cell.
RNA interference
cDNA fragment was amplified using PCR with primers for Gγ1. Primer sequences for RNAi construct were: forward, ATGGACGTAATGTCATCATC; reverse, TTAGAGAACGGTGCAGGACGA. The target sequence is the ORF, which is common to all transcriptional forms of Gγ1 gene. The PCR product was cloned by TA cloning kit (Invitrogen, CA, USA) and sequenced. An inverted-repeat transgene for Gγ1 in inducible Gal4 element was constructed and supplied by the Genetic Strains Research Center, Invertebrate Genetics Laboratory, National Institute of Genetics (Mishima, Japan).
Quantitative PCR
QPCR was performed on cDNA prepared from labella using Brilliant SYBR Green QPCR Master Mix reagents (Stratagene, California, USA) and the thermal cycler apparatus from Stratagene Mx3000P, according to the manufacturer's recommendations. Total cDNA was prepared following the RT–PCR method, except using poly dT24 primer. Primer sequences were designed to synthesize cDNA sized to 216 bp for Gγ1 (forward, ATGGACGTAATGTCATCATC; reverse, TTAGAGAACGGTGCAGGACGA). Elongation factor 1, Ef1α48D (CG8280; 229 bp, forward, CCAACATGGGCAAGGAAAAG; reverse, ATCGATGGTGATACCACGCT), was used to normalize cellular mRNA contents of every preparation. Reactions were performed with 10 μl of enzyme mix, 10 pmol of forward primers, 10 pmol of reverse primers and 1 μl of diluted cDNAs in a final volume of 20 μl. PCR running was performed as follows: initial denaturation at 95°C for 5 s, 40 amplification cycles including annealing, elongation and real-time fluorescence measurement at 55°C for 15 s and denaturation at 95°C for 1 min. At the end of the 40 PCR cycles, the melting temperature was determined by continuously recording the fluorescence during progressive heating up to 95°C with a ramp rate of 0.1°C/s. Three duplicate reaction mixtures were averaged in each PCR run. We performed QPCR at least three times for each strain.
Supplementary Material
Figure S1
Figure S2
Acknowledgments
We thank W Benjamin, C O'Kane, T Kitamoto, F Mastuzaki and the Drosophila Genetic Resource Center in Kyoto Institute of Technology and the Bloomington Stock Center for providing fly strains. We thank P Lucas, N Meunier and F Marion-Poll for helpful suggestions and discussions. We also thank F Yokohari (Fukuoka University) for the use of confocal microscope, K Kimura and M Haruta for technical assistance. HI was supported by the Japan Society for the Promotion of Science. Financial support for this work was provided by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan to TT and HI.
Author contributions: HI and TT designed the research; HI performed the research; HI and TT analyzed the data; KT and RU contributed new analytic tools, and HI and TT wrote the paper.
References
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR (2000) Candidate taste receptors in Drosophila. Science 287: 1830–1834 [DOI] [PubMed] [Google Scholar]
- Duffy JB, Harrison DA, Perrimon N (1998) Identifying loci required for follicular patterning using directed mosaics. Development 125: 2263–2271 [DOI] [PubMed] [Google Scholar]
- Dunipace L, Meister S, McNealy C, Amrein H (2001) Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr Biol 11: 822–835 [DOI] [PubMed] [Google Scholar]
- FlyBase Gene Ontology Report, http://flybasenet/fbservlet/goreport?id=GO:0005834command=linkslink=GNF [Google Scholar]
- Hardie RC, Raghu P (2001) Visual transduction in Drosophila. Nature 413: 186–193 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, Ueda R, Uemura T, Yoshihara M, Goto S (2002) GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis 34: 58–61 [DOI] [PubMed] [Google Scholar]
- He W, Yasumatsu K, Varadarajan V, Yamada A, Lem J, Ninomiya Y, Margolskee RF, Damak S (2004) Umami taste responses are mediated by alpha-transducin and alpha-gustducin. J Neurosci 24: 7674–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi M, Marion-Poll F, Tanimura T (2002) Differentiated response to sugars among labellar chemosensilla in Drosophila. Zool Sci 19: 1009–1018 [DOI] [PubMed] [Google Scholar]
- Hiroi M, Meunier N, Marion-Poll F, Tanimura T (2004) Two antagonistic gustatory receptor neurons responding to sweet–salty and bitter taste in Drosophila. J Neurobiol 61: 333–342 [DOI] [PubMed] [Google Scholar]
- Hodgson ES, Lettvin JY, Roeder KD (1955) Physiology of a primary chemoreceptor unit. Science 122: 417–418 [DOI] [PubMed] [Google Scholar]
- Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJP, Zuker CS (1999) Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96: 541–551 [DOI] [PubMed] [Google Scholar]
- Huang LQ, Shanker YG, Dubauskaite J, Zheng JZ, Yan WT, Rosenzweig S, Spielman AI, Max M, Margolskee RF (1999) G gamma 13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat Neurosci 2: 1055–1062 [DOI] [PubMed] [Google Scholar]
- Ishimoto H, Tanimura T (2004) Molecular neurophysiology of taste in Drosophila. Cell Mol Life Sci 61: 10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Itoh-Furuya A, Fuse N, Matsuzaki F (2004) Differential functions of G protein and Baz-aPKC signaling pathways in Drosophila neuroblast asymmetric division. J Cell Biol 164: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Shimozawa T, Tanimura T (1986) Isolation of Drosophila mutants with abnormal proboscis extension reflex. J Exp Zool 239: 393–399 [DOI] [PubMed] [Google Scholar]
- Kitamoto T (2001) Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol 47: 81–92 [DOI] [PubMed] [Google Scholar]
- Koganezawa M, Shimada I (2002) Inositol 1, 4, 5-trisphosphate transduction cascade in taste reception of the fleshfly, Boettcherisca peregrina. J Neurobiol 51: 66–83 [DOI] [PubMed] [Google Scholar]
- Li XD, Staszewski L, Xu H, Durick K, Zoller M, Adler E (2002) Human receptors for sweet and umami taste. Proc Natl Acad Sci USA 99: 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann B (2001) Receptors and transduction in taste. Nature 413: 219–225 [DOI] [PubMed] [Google Scholar]
- Mclaughlin SK, Mckinnon PJ, Margolskee RF (1992) Gustducin is a taste-cell-specific G-protein closely related to the transducins. Nature 357: 563–569 [DOI] [PubMed] [Google Scholar]
- Meunier N, Marion-Poll F, Rospars JP, Tanimura T (2003) Peripheral coding of bitter taste in Drosophila. J Neurobiol 56: 139–152 [DOI] [PubMed] [Google Scholar]
- Murakami M, Kijima H (2000) Transduction ion channels directly gated by sugars on the insect taste cell. J Gen Physiol 115: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng LX, Zhao G, Ryba NJP, Zuker CS (2002) An amino-acid taste receptor. Nature 416: 199–202 [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang YF, Ryba NJP, Zuker CS (2001) Mammalian sweet taste receptors. Cell 106: 381–390 [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H (2001) A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA 98: 12596–12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Ganguly R (1992) The Drosophila G-protein gamma-subunit gene (d-G-gamma-1) produces 3 developmentally regulated transcripts and is predominantly expressed in the central-nervous-system. J Biol Chem 267: 6086–6092 [PubMed] [Google Scholar]
- Ray K, Ganguly R (1994) Organization and expression of the Drosophila melanogaster G-protein gamma-subunit gene. FASEB J 8: A1399. [DOI] [PubMed] [Google Scholar]
- Rodrigues V, Siddiqi O (1981) A gustatory mutant of Drosophila defective in pyranose receptors. Mol Gen Genet 181: 406–408 [DOI] [PubMed] [Google Scholar]
- Schulz S, Huber A, Schwab K, Paulsen R (1999) A novel G gamma isolated from Drosophila constitutes a visual G protein gamma subunit of the fly compound eye. J Biol Chem 274: 37605–37610 [DOI] [PubMed] [Google Scholar]
- Scott K, Brady R Jr, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R (2001) A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104: 661–673 [DOI] [PubMed] [Google Scholar]
- Sweeney ST, Broadie K, Keane J, Niemann H, O'Kane CJ (1995) Targeted expression of tetanus toxin light-chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14: 341–351 [DOI] [PubMed] [Google Scholar]
- Talluri S, Bhatt A, Smith DP (1995) Identification of a Drosophila G-protein alpha-subunit (dG(Q)alpha-3) expressed in chemosensory cells and central neurons. Proc Natl Acad Sci USA 92: 11475–11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura T, Isono K, Takamura T, Shimada I (1982) Genetic dimorphism in the taste sensitivity to trehalose in Drosophila melanogaster. J Comp Physiol 147: 433–437 [Google Scholar]
- Tanimura T, Shimada I (1981) Multiple receptor proteins for sweet taste in Drosophila discriminated by papain treatment. J Comp Physiol 141: 265–269 [Google Scholar]
- White BH, Osterwalder TP, Yoon KS, Joiner WJ, Whim MD, Kaczmarek LK, Keshishian H (2001) Targeted attenuation of electrical activity in Drosophila using a genetically modified K+ channel. Neuron 31: 699–711 [DOI] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF (1996) Transduction of bitter and sweet taste by gustducin. Nature 381: 796–800 [DOI] [PubMed] [Google Scholar]
- Zhang YF, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu DQ, Zuker CS, Ryba NJP (2003) Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112: 293–301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2


