Abstract
Alternative sources such as buccal cells have already been tested for genetic studies and epidemiological investigations. Thirty-seven volunteers participated in this study to compare cytology brushes, mouthwash, and treated cards for DNA collection. Quantity and quality of DNA and cost and feasibility were assessed. The mean DNA yield at 260 nm was found to be 3.5, 4, and 2.6 μg for cytobrushes, mouthwashes, and treated cards, respectively. A second quantification technique by fluorescence showed differences in the DNA yield with 1.1 and 5.2 μg for cytobrushes and mouthwash, respectively. All buccal samples allowed isolation of DNA suitable for polymerase chain reaction. According to the procedure of sample collection, the yield and purity of collected DNA, and storage conditions, the use of cytobrush appears to be the more appropriate method for DNA collection. This protocol has been validated and is currently applied in three large-scale multicentric studies including adults or children.
INTRODUCTION
In order to establish a DNA bank from a large number of epidemiological studies, we needed to develop collection methods suitable for young subjects as well as for adults using a simple protocol, especially applicable in the case of multicentric studies. Biological samples such as buccal cells can supply DNA for genetic testing and provide a noninvasive approach. Different types of buccal swabs and mouthwash or treated cards have been used to obtain DNA suitable for PCR amplification [1, 2, 3, 4, 5, 6]. The difficulty of obtaining blood samples in order to establish a DNA bank prompted us to compare different methods for buccal cells collection in a preliminary study (consisting of 37 volunteers) which could be subsequently adapted on a large scale. Different protocols to obtain genomic DNA from buccal cells were evaluated: cytobrushes, mouthwash, or treated cards (IsoCode Stix Schleicher & Schuell, Dassel, Germany), to determine the most reliable and cost-efficient method. The amount and quality of DNA and the influence of a lag time at room temperature, to simulate delays of sample mailing, were analyzed in parallel to a cost study. The same protocol for DNA extraction was used and two methods for DNA quantification were compared. Our results confirmed that the protocol based on cytobrushes should be preferentially used, because of its self-collection potential and good quality and sufficient quantity of DNA. This protocol has been validated and currently applied in three large-scale studies including adults or children.
MATERIALS AND METHODS
Preliminary study
Study population
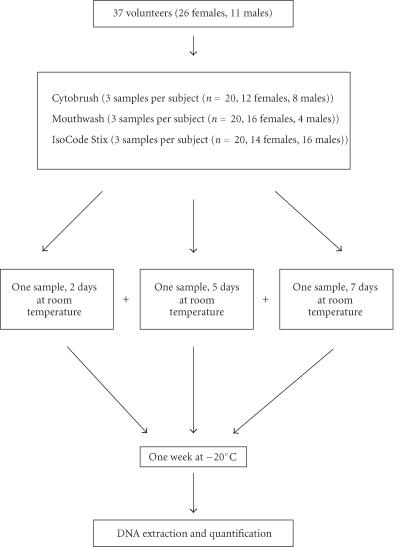
A group of 37 volunteers (26 females, 11 males, with an age range of 20–54 years) was asked to collect buccal cells according to three different methods. Twenty subjects were included in each protocol. Six participated in all the three protocols, 5 in the two protocols, cytobrushes and mouthwash, 3 in cytobrushes and treated cards, and 3 subjects in mouthwash and treated cards. Twenty subjects participated in only one protocol: 6 for cytobrush, 6 for mouthwash, and 8 for treated cards. The time between the three different methods of collection was up to one month. The samples were totally anonymous with no connection between results and volunteer identity.
Sample collection
Three methods of self-collection were compared. The subjects were asked to refrain from smoking, drinking, or eating 45 minutes before sample collection. The first method was to collect cells on a sterile cytobrush (Histobrush, Hardwood Products Company, USA) by twirling it on the inner cheek for 15 seconds. The operation was repeated three times for each subject, on the two cheeks. The swabs were separated from the stick with scissors and transferred to a cryotube. In the second method, buccal cells were collected by rinsing the mouth for 10 seconds with 10 mL of sterile water and expectorating the rinse in a 50- mL centrifuge tube [4] (TPP, Switzerland). This operation was repeated three times for each subject. The third method used is based on IsoCode Stix. These cards were treated to inhibit the growth of bacteria and kill viruses. The subjects expectorated saliva into a sterile cup. The tip of the treated card triangle was placed into the saliva, which wicked onto the matrix. The treated card was air-dried and placed in a bag with desiccant.
Sample processing
Each of the three samples was stored at room temperature for two, five, and seven days, to simulate the mail delay, followed by one week at −20°C before the extraction step (Figure 1).
Figure 1.
Diagram of sample processing. For each method of buccal cell collection, three samples were stored for 2, 5, or 7 days at room temperature to simulate the mail delay before one week at −20°C and DNA extraction.
DNA extraction
DNA was extracted according to the protocol of the manufacturer (QIAamp DNA blood MiniKit, Qiagen, Courtaboeuf, France) and kept at −20°C until use.
For the cytobrush protocol, 400 μL PBS were added to the cytobrush in the cryotube. After vortexing for 15 seconds and incubating at 56°C for 15 minutes 400 μL ethanol (96%–100%) were added and mixed by vortexing. For the mouthwash protocol, cells were pelleted by centrifuging the 50- mL tube for 10 minutes at 1 500 g. The pellet was resuspended in 200 μL of supernatant. The DNA was eluted with 150 μL buffer AE into a clean 1.5 mL microfuge tube for cytobrushes and 200 μL for mouthwash. For treated card protocol, DNA was extracted according to the protocol of the manufacturer (Schleicher & Schuell) and template for amplification was contained in the 100 μL eluate [3].
DNA quantification
DNA concentration was determined at 260 nm using a Power Wave X spectrophotometer (Bio-Tek instruments, inc, Saint-Quentin en Yvelines, France). The ratio 260/280 was measured to evaluate DNA purity. All samples of the preliminary study were analyzed by spectrometry. The second method using the PicoGreen reagent (Interchim, Montluçon, France) was evaluated on a panel of 10 DNA samples from cytobrushes and mouthwashes after 5 days' storage at room temperature (corresponding to the mean delay before the treatment in the laboratory). At the beginning of the preliminary study we did not use the PicoGreen in the laboratory. Several months later we developed this technique and decided to compare the two methods by using the new panel of 10 samples. PicoGreen quantification reagent is able to quantify 25 pg/mL of DNA with a standard spectrofluorometer. The bacteriophage lambda DNA was used to prepare a standard curve (0.05 ng/μL–1.4 ng/μL). Ten microliters, for each point of the standard curve, were added in the 96-well microplate (Costar, VWR, Strasbourg, France), and 2 μL of each sample, completed to 50 μL with 1 mM Tris, 10 mM EDTA, and 50 μL of the PicoGreen dilution were added in each well. The standard curve and the samples were excited at 485 nm and the fluorescence emission intensity was measured at 530 nm, using an FL600 microplate fluorescence reader (Bio-Tek instruments, Inc).
DNA quality
The total DNA size was observed on a 0.8% agarose gel (migration for 2 hour at 100 V) using 5 μL of total DNA and 2 μL of loading buffer.
PCR assays
DNA integrity was evaluated by PCR analysis. First, for each sample exon 3 of the myeloperoxidase gene was amplified on a thermocycler Perkin Elmer 9600. The expected size of amplified fragment corresponds to 481base pairs (bp) [7]. Second, a long PCR was performed on five DNA samples from each protocol. A 5kilobase (kb) fragment was amplified using specific primers of the cytochrome P450 2D6 gene [8]. One-third of the reaction was analyzed on a 1.5% agarose gel.
PCR-RFLP
Five DNA samples obtained by all three protocols for buccal cell collection were assayed by the PCR-RFLP method for the polymorphism in the glutathione S-transferase P1 gene. The primers amplify a 176- bp fragment [9]. The restriction fragments generated with the Bsma1 enzyme (Biolabs, Ozyme, France) digestion are 91 and 85 bp for the variant allele and 176 bp for the wild-type allele (undigested fragment). The results were analyzed on 8% polyacrylamide gel.
Validation of the buccal cell protocol with cytobrushes on a large scale
Three different epidemiological investigations (referred to as Lymphome, Escale, Icare) currently conducted in France were used to validate the protocol based on cytobrush collection, the DNA extraction described above, and the PicoGreen quantification. These collections are stored in the biological resource center located in the INSERM unit U490 (University of René Descartes, 45 rue des Saints-Peres, 75270 Paris Cedex 06, France). Lymphome is a study of genetic and environmental factors in adult lymphoma and 89 subjects (mean age: 53.4 years; range: 20–74) were analyzed; Escale is a study of children leukemia and 93 controls (mean age: 6.75 years; range: 0.07–14.77) were analyzed. The cytobrushes used in these two studies are presented in a hard pack (Master Buccal swab, Tebu International, Le Perray en Yvelines, France). The subjects sent four cytobrushes by mail after self-collection with (for Lymphome) or without (for Escale) the presence of an interviewer. The swabs were then separated from the stick with scissors and transferred into a cryotube in the laboratory before storage at −80°C. Icare is designed to study adult aerodigestive cancers and 180 subjects (mean age: 57.7 years; range: 30.3–75.3) were analyzed. Each subject used four cytobrushes in a soft pack (Histobrush) for self-collection of buccal cells. The swabs were then separated from the stick with scissors and transferred to a cryotube immediately after collection by the interviewer. Finally, the cryotubes were sent by mail to the laboratory and stored at −80°C.
Cost and time of treatment study
For each protocol, the different steps of collection were evaluated to establish the final cost by sample, including tubes, reagents, mailing, DNA extraction, storage materials, and the time of preparation before storage.
RESULTS
Total DNA yield and quality
Means of total DNA yield measured at 260 nm from the three collection methods are presented in Table 1, and the average amount of DNA ranged from 0.4 to 9 μg per cytobrush, 0.6 to 21.7 per mouthwash, and 0.4 to 9.3 per triangle of treated card. The greater total DNA yield was obtained from mouthwash and cytobrush (4 μg and 3.5 μg, resp). One triangle of treated card gave less DNA (2.6 μg). An effect of lag time before extraction was observed for mouthwash, the amount of extracted DNA decreased by 50% between samples kept for 2 days or 7 days at room temperature. For the two other methods of collection there was no significant decrease of DNA yield between 2, 5, or 7 days (Table 1). The quantification at 260 nm and with PicoGreen for the panel of 10 samples is presented in Table 2. The values obtained with PicoGreen show a decrease of 2 for cytobrush and 1.25 for mouthwash with higher variations for mouthwash (SD = 4.4 at 260 nm and 3.8 with PicoGreen instead of 1.4 and 1.2 for cytobrush). The values of the ratio 260/280 are presented in Table 1. The mean value is 1.1 for IsoCode, 1.1 for cytobrush, and 1.7 for mouthwash. It appears that for treated card the purification let proteins in the solution of extracted DNA. DNA was evaluated firstly by gel electrophoresis. For cytobrush and mouthwash, a visible band migrated at 23 kb, indicating the presence of high molecular weight DNA, but a smear over a broad size range was also observed. No visible DNA appears on the agarose gel for treated card (data not shown). Then, DNA samples were tested for amplification by using different PCR protocols. For mouthwash and cytobrush (n = 60 for each procedure; Figure 1) 98.3% of samples (n = 59) gave amplification for the 481 bp for exon 3 of myeloperoxidase gene and 96.6% (n = 58) of the samples for treated card gave a positive signal. Among 5 samples from each protocol, only 3 collected by mouthwash and 2 collected by cytobrush gave amplification with the “long-range” PCR test; no DNA extracted from treated card amplified the 5 kb fragment. The ability to use these DNA for the detection of mutation with PCR-RFLP was tested for the polymorphism in the glutathione S-transferase P1 gene. The five DNA samples tested for cytobrush and mouthwash were usable in this method but the restriction analysis was not discriminating for DNA extracted from treated card, in part due to low PCR signal (data not shown).
Table 1.
Total DNA yield and range (in parentheses) in μg three times, 20 subjects for the three different methods of buccal cell collection. The amount of DNA was measured on a spectrophotometer at 260 nm and at 280 nm to determine the ratio 260/280.
| Time for storage at room temperature | Cytobrush | Mouthwash | Triangle of treated card |
| before a week at −20°C | N = 20 | N = 20 | N = 20 |
| 2 days | 3.4 | 5.1 | 2.9 |
| (0.4–8.5) | (1.6–21.7) | (1.0–9.3) | |
| 5 days | 3.5 | 4.3 | 2.9 |
| (1.4–6.2) | (0.4–8.1) | (0.4–7.9) | |
| 7 days | 3.5 | 2.5 | 2.4 |
| (0.9–9) | (0.6–5.2) | (0.7–5.6) | |
| Mean of total DNA yield (μg) | 3.5 ± 0.1 | 4 ± 1.3 | 2.6 ± 0.3 |
| Mean ratio 260/280 | 1.6 ± 0.2 | 1.7 ± 0.2 | 1.1 ± 0.1 |
Table 2.
Comparison of DNA amounts in μg, obtained from 10 subjects with two different methods of quantification (absorbance at 260 nm or fluorescence protocol with PicoGreen).
| Mean total DNA yield (μg) | Cytobrush | Mouthwash |
| N = 10 | N = 10 | |
| Spectrophotometer (260 nm) | 2.4 ± 1.4 | 6.5 ± 4.4 |
| (0.9–5.7) | (2.7–16.5) | |
| PicoGreen | 1.1 ± 1.2 | 5.2 ± 3.8 |
| (0.04–4.04) | (2.2–12.4) | |
The data obtained for the three current epidemiological studies are presented in Table 3 and are similar to those obtained with the panel (Table 2). No difference was observed related to the origin of the samples (ie, children or adult leading to 1.2 and 1.3 μg, resp.) or the presentation of cytobrushes (hard pack or soft pack, 1.3 μg each). The intervention of an interviewer did not influence the final quantity of DNA (with or without an interviewer, 1.3 and 1.2 μg, resp).
Table 3.
Total DNA in μg from three epidemiological studies obtained with the protocols Escale and Lymphome correspond to self-collection in a hard pack by children in the absence of an interviewer (Escale) or correspond to self-collection in a hard pack in the presence of an interviewer (Lymphome). The protocol Icare allowed self-collection in adults in the presence of an interviewer. DNA was extracted according to QIAamp DNA blood MiniKit, QIAGEN, Courtaboeuf, France. The quantity of DNA was measured by fluorescence with PicoGreen.
| Escale | Lymphome | Icare | |
| (n = 93) | (n = 89) | (n = 180) | |
| Mean total DNA yield (μg) | 1.2 ± 1.1 | 1.3 ± 1.3 | 1.3 ± 1.7 |
| (0.1–7.3) | (0.04–5.6) | (0.03–17.7) | |
DISCUSSION
The aim of our study is to evaluate different protocols for DNA collection from buccal cells, to develop a simple method for collection from children and adult control subjects. Three methods (cytobrush, mouthwash, and treated card) were compared and both the quantity and quality of DNA and the feasibility and cost of the procedures were assessed.
We chose to study the utilization of buccal cells because it is a noninvasive method, which allows many genotyping. Other samples can be used for DNA banking or genotyping: blood sampling is an invasive method and must be done by a medical person; urines also provide DNA but with variable yields and contain PCR inhibitors. Our results showed that the buccal cells provided usable amounts of DNA, particularly from mouthwash and cytobrush. For a cytobrush sample, the amount of extracted DNA gave interindividual variation depending on the buccal mucosa and the force exercised with the brush. We previously checked that two consecutive brushes did not affect the DNA yield (data not shown). The treated card corresponded to the lowest efficiency because of the small quantity of collected saliva. Previous studies reported the greatest efficiency for DNA extraction from mouthwash but similar values for cytobrush, based on the same DNA extraction method [2, 10]. The DNA yield of collection may depend on the collection protocol which may include a cheek and tongue brushing before mouthwash [1, 4, 11] or rubbing the cheeks against teeth to prepare the buccal mucosa [10]. The duration of collection can influence the DNA yield. In the present study, the time of brushing corresponded to 15 seconds for cytobrush and 10 seconds for mouthwash. Other studies preferably used 30 seconds for cytobrush and 60 seconds for mouthwash in order to increase the amount of DNA [1, 2, 10, 11] but these conditions appear very difficultly compatible with studies including children. In the preliminary study, two methods of DNA quantification were tested on 10 samples stored for five days at room temperature and one week at −20°C. The fluorescence based on the reagent PicoGreen, which allows the determination of very small DNA concentration, is more precise than the measurement at 260 and 280 nm, because of its specificity for double-stranded DNA (dsDNA). The high specificity of PicoGreen for dsDNA can explain the difference observed when DNA quantification was performed with the measurement at 260 nm. The presence of proteins or other contaminants could lead to the erroneous results obtained with the spectrometric method and strongly suggest that PicoGreen is an alternative protocol more suitable for specific dsDNA quantification, using a very small volume of extracted DNA (2 μL of sample instead of 100 μL for spectrometric method). The quality of DNA assessed by the measurement at 280 nm showed some impurities according to the different protocols. Especially with extracted DNA from treated card which displayed a ratio 260/280 of 1.1, the purification let some proteins in the solution of extracted DNA, so the DNA can not be kept for long-term conservation (several years in the case of DNA banking). The measurement at 260 and 280 nm could be modified by RNA, single-stranded DNA, or degraded DNA but PicoGreen could access the really available amount of DNA for PCR analysis. Previous studies and our results demonstrated that the PicoGreen-based assay has excellent reproducibility and sensitivity [12]. Furthermore, comparison of spectrometric method and other dye-binding assays (eg, Hoescht reagent) showed that PicoGreen was found to be more sensitive and suitable for DNA quantification of low sample volume [13, 14]. It is important to manipulate small DNA aliquots instead of the stock solution in order to avoid DNA contamination and to allow high-throughput genotyping. The quality of DNA was then evaluated by PCR and our results showed that the extracted DNA could be amplified for short fragments in most of the cases with the three sampling procedures. Long fragment amplification displays weak efficiency for mouthwash and cytobrush and failed with treated card. After PCR, buccal DNA from cytobrush or mouthwash can be easily used for PCR-RFLP or TaqMan methods for genotyping, as is described elsewhere [5, 11]. Finally, the effect of the lag time of storage at room temperature was visible for mouthwash while the cytobrushes were less sensitive to the lag time at room temperature (stable for at least one week). This could be crucial for multicentric studies, which require mailed samples.
In our study the QIAamp DNA kit for DNA extraction was used, which allowed extracting DNA at high throughput in microplate (QIAamp 96 DNA blood Kit, Qiagen, Courtaboeuf, France) leading to 260/280 ratio comprising between 1.7 and 1.9. In previous studies different methods of DNA extraction have been tested, like phenol-chloroform, Puregene kit (Gentra Systems, Minneapolis, MN), and this could influence the DNA yield and integrity [2].
The cost study (Table 4) showed advantages of treated cards; this method is more economic because of the absence of an extraction step with a commercial kit. It is a rapid method to obtain DNA for diagnosis of genetic diseases but cannot be adapted to epidemiological studies and genotyping on a large scale. The mouthwash is more expensive than cytobrush; the weight for one sample collected by mouthwash (in 10 mL buffer) is higher than a cytobrush sample and increases the cost of mailing. The time of treatment before storage should also be taken into account (Table 4): for mouthwash, it is three times that of cytobrush or treated card because of the need for centrifugation.
table 4.
Time of treatment before storage in minutes. Final cost (Euros) per sample according to the different steps of collection.
| Cytobrush | Mouthwash | Treated card | |
| (n = 1) | (n = 1) | (4 triangles)* | |
| Time of treatment before storage (min) | 5 | 15 | 5 |
| Cost before storage | 1.1 | 3.5 | 1.1 |
| Collection | 1.3 | 0.54 | 0.82 |
| Mail1 | 0.46 | 1.02 | 0.46 |
| Storage | 0.1 | 0.41 | 0.41 |
| Extraction | 1.7 | 1.7 | 0.025 |
| Final cost (Euros HT) | 4.66 | 7.17 | 2.81 |
*Only available under a format of 4 triangles.
1“Mail” corresponds to the postage rates in France. The storage procedure includes a well of a cryobox (Nalgène system 100, VWR, Strasbourg, France) for each protocol plus a cryotube (Greiner, 2 mL, VWR, Strasbourg, France) for mouthwash and treated card at −80°C. The DNA extraction method used is QIAamp DNA blood MiniKit (Qiagen) for cytobrush and mouthwash and IsoCode Stix (Schleicher & Schuell) for treated card.
Although buccal cells gave a smaller amount of DNA than blood, recently developed methods of genotyping use very small amounts of DNA (2–10 ng per assay) and thus allow the use of buccal cells as a source of DNA. Concerning the buccal cells, the short time of collection (15 seconds for cytobrush and 10 seconds for mouthwash) and the fact that the subject can perform it themselves without the operation of a physician or a nurse were essential for collection in multicentric studies and mail delivery. We did not evaluate the potential presence of bacterial DNA in these buccal samples. Garcia-Closas et al. [2] reported that human DNA for cytobrush was lower than that for mouthwash and suggested the use of an alcohol-containing mouthwash, which is not appropriate for children and self-collection. Even if cytobrush and mouthwash provided a correct amount of DNA, mouthwash is more distasteful, not applicable with young people, and more expensive in large-scale studies. Although the mouthwash displayed the highest efficiency for DNA amount, the need of cells in 10- mL suspension for transport before extraction constituted a serious disadvantage for multicentric studies. We finally demonstrated that cytobrushes could be used for studies with very young subjects on a large scale (Table 3) [11] and in the absence of an interviewer (Table 3). In conclusion, the use of cytobrush appears to be the most appropriate method to facilitate the self-collection of human genomic DNA with good quality and high security particularly in multicentric studies and to give good value for money for DNA banking.
ACKNOWLEDGMENT
The authors thank Claudia de Toma for helping with fluorescent quantification. This work is supported by La Ligue Nationale Contre le Cancer (Comité de Paris), the Association pour la Recherche Contre le Cancer, and the Conseil Régional d'Ile-de-France.
References
- 1.Feigelson H.S, Rodriguez C, Robertson A.S, et al. Determinants of DNA yield and quality from buccal cell samples collected with mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10(9):1005–1008. [PubMed] [Google Scholar]
- 2.Garcia-Closas M, Egan K.M, Abruzzo J, et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10(6):687–696. [PubMed] [Google Scholar]
- 3.Harty L.C, Garcia-Closas M, Rothman N, Reid Y.A, Tucker M.A, Hartge P. Collection of buccal cell DNA using treated cards. Cancer Epidemiol Biomarkers Prev. 2000;9(5):501–506. [PubMed] [Google Scholar]
- 4.Harty L.C, Shields P.G, Winn D.M, Caporaso N.E, Hayes R.B. Self-collection of oral epithelial cell DNA under instruction from epidemiologic interviewers. Am J Epidemiol. 2000;151(2):199–205. doi: 10.1093/oxfordjournals.aje.a010188. [DOI] [PubMed] [Google Scholar]
- 5.Heath E.M, Morken N.W, Campbell K.A, Tkach D, Boyd E.A, Strom D.A. Use of buccal cells collected in mouthwash as a source of DNA for clinical testing. Arch Pathol Lab Med. 2001;125(1):127–133. doi: 10.5858/2001-125-0127-UOBCCI. [DOI] [PubMed] [Google Scholar]
- 6.Walker A.H, Najarian D, White D.L, Jaffe J.F, Kanetsky P.A, Rebbeck T.R. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect. 1999;107(7):517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevrier I, Stucker I, Houllier A.M, et al. Myeloperoxidase: new polymorphisms and relation with lung cancer risk. Pharmacogenetics. 2003;13(12):729–739. doi: 10.1097/01.fpc.0000054143.14659.ba. [DOI] [PubMed] [Google Scholar]
- 8.Johansson I, Lundqvist E, Dahl M.L, Ingelman-Sundberg M. PCR-based genotyping for duplicated and deleted CYP2D6 genes. Pharmacogenetics. 1996;6(4):351–355. doi: 10.1097/00008571-199608000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Harris M.J, Coggan M, Langton L, Wilson S.R, Board P.G. Polymorphism of the Pi class glutathione S-transferase in normal populations and cancer patients. Pharmacogenetics. 1998;8(1):27–31. doi: 10.1097/00008571-199802000-00004. [DOI] [PubMed] [Google Scholar]
- 10.King I.B, Satia-Abouta J, Thornquist M.D, et al. Buccal cell DNA yield, quality, and collection costs: comparison of methods for large-scale studies. Cancer Epidemiol Biomarkers Prev. 2002;11(10 pt 1):1130–1133. [PubMed] [Google Scholar]
- 11.London S.J, Xia J, Lehman T.A, et al. Collection of buccal cell DNA in seventh-grade children using water and a toothbrush. Cancer Epidemiol Biomarkers Prev. 2001;10(11):1227–1230. [PubMed] [Google Scholar]
- 12.Ahn S.J, Costa J, Emanuel J.R. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996;24(13):2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami P, McCaman M.T. Quantitation of adenovirus DNA and virus particles with the PicoGreen fluorescent Dye. Anal Biochem. 1999;274(2):283–288. doi: 10.1006/abio.1999.4282. [DOI] [PubMed] [Google Scholar]
- 14.Singer V.L, Jones L.J, Yue S.T, Haugland R.P. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem. 1997;249(2):228–238. doi: 10.1006/abio.1997.2177. [DOI] [PubMed] [Google Scholar]