Abstract
To catalogue data on chromosomal aberrations in cancer derived from emerging molecular cytogenetic techniques and to integrate these data with genome maps, we have established two resources, the NCI and NCBI SKY/M-FISH & CGH Database, and the Cancer Chromosomes database. The goal of the former is to allow investigators to submit and analyze clinical and research cytogenetic data. It contains a karyotype parser tool, which automatically converts the ISCN short-form karyotype into an internal representation displayed in detailed form and as a colored ideogram with band overlay, and also contains a tool to compare CGH profiles from multiple cases. The Cancer Chromosomes database integrates the SKY/M-FISH & CGH Database with the Mitelman Database of Chromosome Aberrations in Cancer, and the Recurrent Chromosome Aberrations in Cancer database. These three datasets can now be searched seamlessly by use of the Entrez search and retrieval system for chromosome aberrations, clinical data, and reference citations. Common diagnoses, anatomic sites, chromosome breakpoints, junctions, numerical and structural abnormalities, and bands gained and lost among selected cases can be compared by use of the “similarity” report. Because the model used for CGH data is a subset of the karyotype data, it is now possible to examine the similarities between CGH results and karyotypes directly. All chromosomal bands are directly linked to the Entrez Map Viewer database, providing integration of cytogenetic data with the sequence assembly. These resources, developed as a part of the Cancer Chromosome Aberration Project (CCAP) initiative, aid the search for new cancer-associated genes and foster insights into the causes and consequences of genetic alterations in cancer.
INTRODUCTION
The relevance of chromosomal abnormalities to the development of all forms of human malignancies is widely recognized, and the field of cytogenetic oncology plays an important role in cancer research today. To facilitate comprehensive identification of chromosomal aberrations, two complementary, FISH-based molecular cytogenetic techniques were developed in the 1990s, comparative genomic hybridization (CGH; (Kallioniemi et al., 1992) and spectral karyotyping/multiplex-FISH (SKY/M-FISH; Liyanage et al., 1996; Schröck et al., 1996; Speicher et al., 1996). Together with the standard FISH methodologies developed a decade earlier, they have greatly enhanced karyotype interpretation, especially as it pertains to cancer.
The need for cataloguing cytogenetic aberrations in cancer in a systematic, concise, and uniform manner has long been recognized, and the first such effort was published by Mitelman more than 20 years ago. That catalog is now online as the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman) and is the largest online cytogenetics database available (Mitelman et al., 2003). It contains the complete karyotypes, certain patient characteristics, and reference citations from more than 46,000 cases of neoplastic disorders, manually culled from the literature. It is updated quarterly. Another useful cytogenetic Web site is the Atlas of Genetics and Cytogenetics in Oncology and Haematology (http://www.infobiogen.fr/services/chromcancer/), an online journal and database, that reviews and summarizes data on genes, cytogenetics, and clinical entities in cancer and cancer-prone diseases (Huret et al., 2000). Several online databases display CGH data: Charite (http://amba.charite.de/cgh/); Progenetix (www.progenetix.net/; Baudis and Cleary, 2001); the Laboratory of Cytomolecular Genetics (CMG), Helsinki, Finland (http://www.helsinki.fi/cmg/); and the CGH Data Base, Tokyo (http://www.cghtmd.jp/cghdatabase/index_e.htm); most of these databases display cases from a single laboratory, but the Progenetix database contains CGH profiles for more than 10,000 cases from the literature. Only a few online databases display SKY data: Chromosome Rearrangements in Carcinomas (http://www.path.cam.ac.uk/~pawefish/) and Cell Line NCI60 Drug Discovery Panel (http://home.ncifcrf.gov/CCR/60SKY/new/demo1.asp); these two SKY databases contain the results from only a few studies.
To share and compare results and to link chromosomal aberration with sequence maps, the National Cancer Institute (NCI) and the National Center for Biotechnology Information (NCBI) of the National Institutes of Health (NIH) have established two databases, the NCI and NCBI SKY/M-FISH and CGH Database (http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi) (Knutsen et al., 2002) and its companion, the Entrez Cancer Chromosomes database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cancerchromosomes). These databases were developed as a part of the Cancer Chromosome Aberration Project (CCAP), which “is a Cancer Genome Anatomy Project (CGAP) initiative designed to develop a set of ‘tools’ to define and characterize the distinct chromosomal alterations that are associated with malignant transformation” (http://cgap.nci.nih.gov/). The goal of the SKY/M-FISH & CGH Database is to allow investigators to submit and analyze both clinical and research (e.g., cell lines) SKY/M-FISH and CGH data. This database is currently designed to contain human and mouse data; other genomes, such as that of the rat, will be added in the future. The database is open to all submitters.
Establishment of that database led to the development of the Entrez Cancer Chromosomes search database, which contains and integrates all the data from the SKY/M-FISH & CGH Database, the Mitelman Database of Chromosome Aberrations in Cancer, and the Recurrent Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/RecurrentAberrations) (Fig. 1). With this additional database, it is possible to not only search for cases with the same cytogenetic and clinical features, but also to look at each case in great detail and search for similarities among all cases of interest. By directly linking each chromosome band to the NCBI Map Viewer database (http://www.ncbi.nlm.nih.gov/mapview), integration of the cytogenetic data with the human and mouse genome assembly has now become possible.
Figure 1.
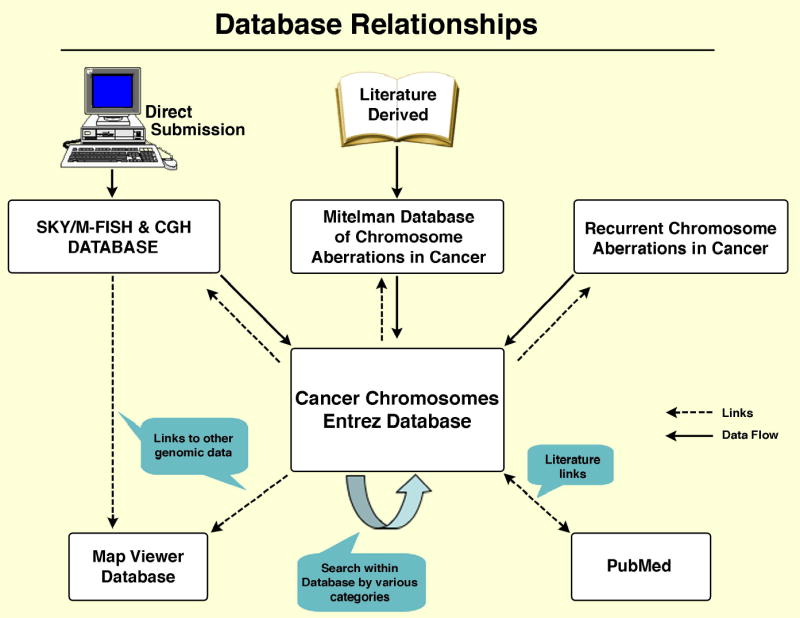
Diagram demonstrating the relationship of the SKY/M-FISH & CGH Database, the NCI Mitelman Database of Chromosome Aberrations, and the NCI Recurrent Chromosome Aberrations in Cancer Database to the Cancer Chromosomes Entrez Database and other databases such as Map Viewer and PubMed. All three databases integrated into Cancer Chromosomes also link directly to PubMed.
DESCRIPTION OF DATABASES
The features and organization of the SKY/M-FISH & CGH Database and the Cancer Chromosomes database are outlined in Table 1. In the following sections, we first describe how to enter data into the SKY/M-FISH & CGH Database, and then demonstrate how to use Cancer Chromosomes to query the data entered into that database as well as in the Mitelman databases.
TABLE 1.
Features and Organization of the Databases
A. SKY/M-FISH & CGH Database
|
| B. Cancer Chromosomes Database |
|
SKY/M-FISH & CGH DATABASE
Data Submission
Registration is required to submit data. All data submitted remain in a “private” mode (i.e., only accessible to the submitter) until the submitter releases it for public viewing within a time period not to exceed two years. Each submitter is responsible for data accuracy. Complete instructions for data entry are included. The submitter enters the name, diagnosis, and site for each case, using the International Classification of Diseases for Oncology, 3rd edition (ICD-O-3; Fritz et al., 2000). To find and select the correct ICD-0-3 morphology and topography terms, the user is referred to the NCI MetathesaurusTM, a medical terminology search engine developed by the NCI (http://ncimeta.nci.nih.gov/indexMetaphrase.html). Mouse diagnosis and site terms are not restricted to the ICD-O-3 terminology. After a unique case number has been assigned, the submitter proceeds to enter the cytogenetic, clinical, and reference data. All terms (such as specific drugs) entered are searchable through the database. All data, whether added manually or automatically, can be edited and updated by the submitter at any time (see below), even after they have been made accessible for public viewing.
For the SKY/M-FISH data entry, the submitter enters the karyotype [for humans, according to the ISCN 1995 nomenclature (ISCN,1995), or, for mice, according to the Rules for Nomenclature of Chromosome Anomalies (Committee on standardized genetic nomenclature for mice, 1996)] and the modal chromosome numbers. Ploidy is then selected from a pull-down menu, followed by either (1) entering the cytogenetic information for each chromosome manually, or (2) using the karyotype parser (see below), which automatically converts the written karyotype into its component elements and from there into an ideogram (the parser is currently available only for human karyotype conversion). For entering the cytogenetic information manually, each chromosome is described from top to bottom, segment by segment, with variation in the amount of each band to be displayed, as shown in Figure 2A. The program permits the drawing of chromosomal aberrations such as ring chromosomes and homogeneously staining regions (hsrs), and variation in the size of unknown segments (unknown band or unknown chromosome) or an hsr as a percentage of the normal parent chromosome (Fig. 2A).
Figure 2.

Building tools: (A) Building an abnormal SKY/M-FISH chromosome. Each chromosome is built from top to bottom by specifying the stop and start bands for each segment; hsrs can be identified by chromosomal origin and size, and ring chromosomes are indicated by a line connecting the top and bottom of the abnormal chromosome. (B) Building an abnormal CGH profile. Each segment of a chromosome showing gain (green) or loss (red) is specified by its stop and start bands and by selecting the appropriate type of gain or loss.
Karyotype Parser
The karyotype parser is a computer program built by one of the authors (R.K., a computational linguist) for automatically reading short-form karyotypes, extracting the intrinsic information, and inserting it into the SKY database. Using the ISCN 1995 rules, the parser (1) breaks the karyotype into small syntactic components, (2) assembles information from these components into an information structure in computer memory, (3) transforms this information into the formats required for an application, and (4) inserts it into the database. The parser works accurately with karyotypes with complete band designations for each breakpoint, but manual editing is required for incomplete karyotypes, questionable identification of a chromosome or chromosome structure, or the presence of hsrs, double minutes (dmin), and so forth.
CGH data can be submitted manually or automatically. If making a manual submission, the submitter enters the band information for each affected chromosome or chromosome segment, working from the top of the chromosome (pter) and describing the start band and stop band for each gain or loss (Fig. 2B); the computer program then displays the final profile. In the automatic format, which is currently possible if the Leica CW4000 and the Applied Imaging Cytovision-High Resolution CGH Genus software (human and mouse) are used, the CGH data can be transferred automatically in order to create the profile from a file download enabled by the software; automatic transfer will also be available soon with the MetaSystems' Isis CGH software.
Data Presentation
The SKY/M-FISH data are displayed in the form of a colored ideogram (termed SKYGRAM) with each normal and abnormal chromosome displayed in its unique classification color, with band overlay. Figure 3 shows a SKYGRAM from OVCAR-8, an ovarian cancer line from the NCI60 Drug Discovery Panel, and Figure 4, a SKYGRAM from a mouse pro-B cell lymphoma. CGH data are displayed on an ideogram with vertical bars indicating gain, loss, and amplification of chromosomal material. Patient clinical information is also displayed, and the literature citation is directly linked to the relevant abstract in PubMed, the NCBI's online retrieval system for biomedical literature abstracts. Each human and mouse chromosome band displayed in the ideogram is automatically linked to the Map Viewer, which integrates map and sequence data from a variety of sources (Dombrowski and Maglott, 2002). For example, by clicking on the breakpoint 17q21, which is prevalent in both breast and ovarian cancer, from the SKYGRAM of another ovarian cancer cell line, SKOV-3, it is possible to retrieve sequence data and information on FISH-mapped BAC clones which can be used to pinpoint the location of specific breakpoints in individual cases (Fig. 5).
Figure 3.
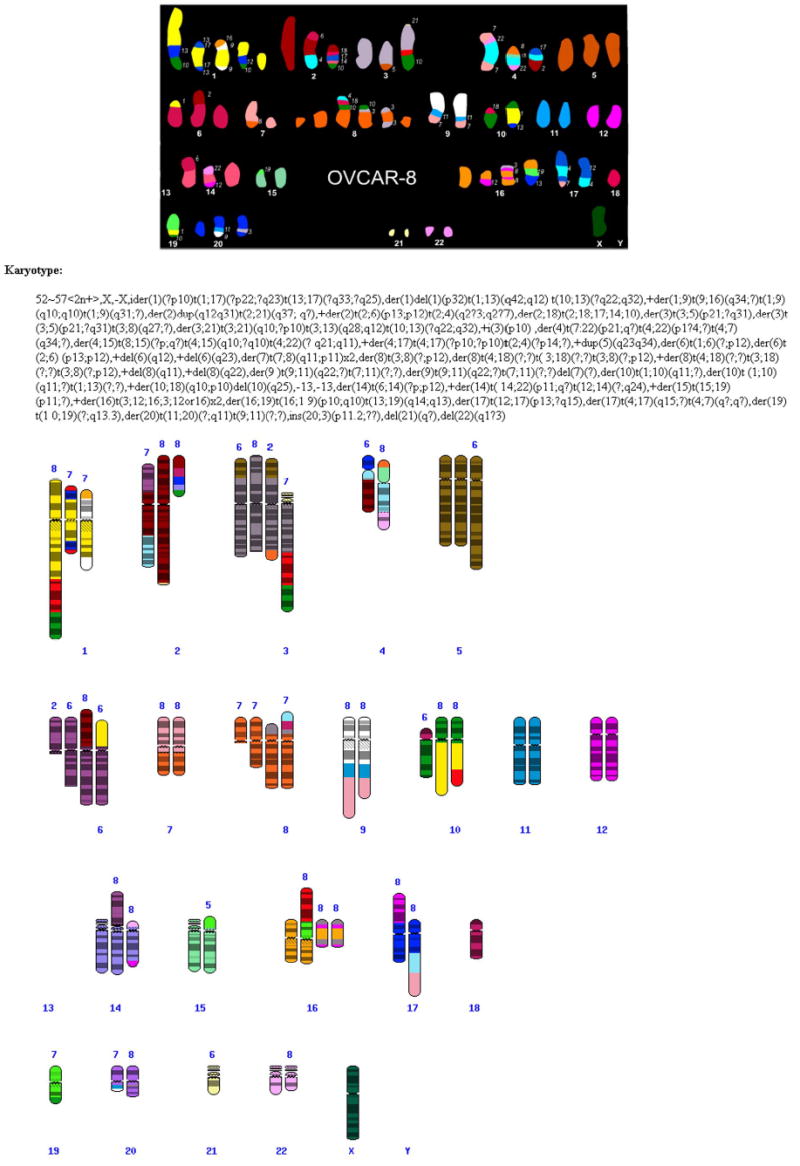
Human cell line OVCAR-8 from the NCI60 Drug Discovery Panel study. Top: SKY classification karyotype from a single cell. Bottom: Complete SKYGRAM based on 8 cells. Chromosome segments without banding indicate that the band origin was unknown.
Figure 4.

Complete SKYGRAM of PKT3, a mouse pro-B cell lymphoma with 12;15 translocations and an hsr composed of alternating sequences from chromosomes 12 (in fuscia) and 15 (in light green), which resulted in amplification of IgH and cMYC.
Figure 5.

Linkage of SKY/M-FISH & CGH cytogenetic data with sequence data via Map Viewer. Left panel: partial SKYGRAM ideogram from SKOV-3, an ovarian cancer cell line, with a 17q21 breakpoint. Clicking on this breakpoint brings up Map Viewer for this band (right panel). Relative alignment and stretching of the sequence maps (Genes_seq, gbDNA, and NCI_Clone) and the cytogenetic maps (Genes_cyto and NCI_FISH_Clones) is done uniformly. The maps labeled "NCI_Clone" and "NCI_FISH_Clone" differ in that the extent and location of the clones, shown by vertical blue lines, are determined by sequence in the former and by observed cytogenetic location in the latter; the gray lines connect the sequence and cytogenetic location for the same clone. Band 17q21 contains the BRCA1 gene, which is involved in both breast and ovarian cancer; the yellow pop-up window provides information about this gene and a link to its sequence.
Because it is useful to compare the CGH profiles of multiple cases, whether they have the same diagnosis or are related in some other way, we have developed the CGH Case Comparison Tool, which displays all of the selected profiles on a single ideogram (Fig. 6A). With this tool, an investigator can select cases on the basis of a variety of criteria. For example, some ovarian cases show deletion of 6q: do these particular cases have other gains or losses in common that would distinguish them from ovarian cases without 6q loss? This can be queried by selecting those cases that have 6q loss and performing another case comparison. This tool can be applied to all public cases in the databases, and, in addition, each submitter can apply it to his own private cases. The display of the cases to be compared can be manipulated in a variety of ways, including bar width, space between bars, display pattern, and legends; each case or group of cases can be displayed in a different color (Fig. 6B). From the resulting ideogram, each individual case profile and case details can be displayed separately.
Figure 6.
CGH Case Comparison tool: (A) A comparison of CGH profiles in 25 cases of human lung cancer and (B) comparison of CGH profiles in mouse mammary tumors with amplification of Her2/neu at 11D: each case is shown in a different color, and the legends at the top of each vertical bar identify the individual cases.
The SKY/M-FISH & CGH Database currently contains more than 1700 cases, 700 of which are available for public viewing (the remaining ones will be released on publication). Data soon to be made public include CGH studies on over 400 cases of different types of lymphoma. SKY and/or CGH data from a wide variety of human leukemias, lymphomas, and solid tumors are available. Mouse data include cytogenetic abnormalities observed in models of solid tumors, leukemias, and lymphomas, and studies of knock-out and transgenic mice. Included among the human cases in the database are 59 cell lines from the NCI60 Drug Discovery Panel (Roschke et al., 2003). The SKY study of these cell lines by Roschke et al. (2003) allowed subclassification of these cell lines based on their structural genome anatomy, offering insights into the causes and consequences of genetic instability that may occur during tumor development and progression.
CANCER CHROMOSOMES DATABASE
The Cancer Chromosomes database further extends the SKY/M-FISH & CGH Database and integrates it with existing databases. We wanted to be able to take the data from that database and perform various manipulations that would track specific abnormalities and relate them in a meaningful way to the sequence-level data. Cancer Chromosomes is part of the Entrez system. Entrez is the text-based search-and-retrieval system used at the NCBI for all of its major databases, including PubMed, Nucleotide and Protein Sequences, OMIM, etc. (Ostell, 2003). Each database within Entrez is a collection of data grouped and indexed together. The system can infer relationships among different data that may suggest future experiments or assist in interpretation of the available information, even though it may come from different sources.
The Cancer Chromosomes integrates data from the SKY/M-FISH & CGH Database, the Mitelman Database of Chromosome Aberrations in Cancer, and the Recurrent Chromosome Aberrations in Cancer database so that they can be searched seamlessly for all chromosome aberrations. Data from the Mitelman databases are added using the karyotype parser, and because it makes the data computationally tractable, it can be used to detect chromosomal gains and losses and directly find breakpoints from the most complex rearrangements. Seamless searching is a process by which all data are searched simultaneously for any particular aberrations. This process employs the use of “pseudo documents,” a conversion of cytogenetic and clinical textual data into one common set of terms for the purposes of statistical comparison (see “Computation of Related Articles” at http://www.ncbi.nlm.nih.gov/entrez/query/static/computation.html). Each term is then assigned a particular weight on the basis of how often it occurs, by use of statistical methods identical to those used in PubMed. Similarities among documents are thus based on words or terms they have in common. Features of Entrez such as History, Clipboard, and Preview/Index function as they do for all Entrez databases. Search tips are provided in the Help document.
Three search formats are offered on the Cancer Chromosomes home page: a conventional Entrez query, a quick/simple search, and an advanced search. The Entrez Query Box is a free text search performed by use of the search box at the top of the page. The simple search offers a set of menus from disease site and diagnosis terms that may be selected and combined with specifications for a particular chromosomal location and anomaly. The advanced search form offers a combination of forms and menus of search terms for complex queries.
Performing Searches
The following example demonstrates how one would go about searching for a particular abnormality and where the results might lead. Search: Which diseases show involvement of chromosome band 14q32 and what other chromosome aberrations are related to 14q32? The initial result is a summary report for each affected case (Fig. 7), with links to the full karyotype, CGH profile, clinical information, and PubMed abstract. One of the cases in the summary report is a CGH study of papillary serous cystadenocarcinoma of the ovary, which shows amplification of 14q32. As in PubMed, there are “Links” to related cases (related karyotypically, by CGH pattern, diagnostically, and/or textually). This particular case was related by CGH to 50 other cases with varying diagnoses in the database, about 40% of which also had 8q24 involvement. For gaining more information on the cases in the summary report, two additional reports can be obtained by use of the pull-down menus in the display bar: a detailed report and a similarity report.
Figure 7.

Cancer Chromosomes Summary Report--all clones/cells in the database involving chromosome band 14q32. Links are provided to each karyotype or CGH profile, case details, and PubMed abstract. The pull-down menu in the Display bar and the Links to the right of each case provide additional tools for comparison of cases. A search based on case information, such as diagnosis and site, result in a case-based report (i.e., a list of all cases displaying the searched element), whereas a search based on underlying cytogenetic features are displayed as a clone/cell report (i.e., each clone or cell is searched separately and is listed separately in the report; in a case with several clones, the report will list only those clones that contain the searched element).
Detailed Report
All details of a case can be viewed by selecting the Detailed Report from the Display pull-down menu. The detailed report provides reference information; the complete karyotype; information on the cytogenetic aspects of each case, such as information on each breakpoint and chromosomal junction (junctions are formed at translocation, inversion, and insertion sites, as well as in the formation of ring chromosomes); the number of structural and numerical aberrations involving each affected chromosome; and all bands gained or lost.
Similarity Report
The result of the search for 14q32 revealed that more than 4000 cases display abnormalities of some sort at this chromosome band. By selecting Similarity Report in the Display pull-down menu, one can compare these cases for a variety of common cytogenetic and clinical elements. For example, as shown in Table 2, the first 500 cases included 109 with follicular lymphoma, and the majority of the remaining cases had other types of lymphoma. In addition to 14q32, the most common breakpoints were 8q24 (122 cases), 11q13, and 18q21. Only the follicular lymphomas (by clicking on the number of cases) could be selected and another similarity report performed. The result would show that 108 of 109 cases listed the site as lymph node, and 105 of 109 involved 18q21, as expected in the t(14;18). It may also be of interest that 12 cases had an i(6p) and that the most common numerical abnormalities were loss of chromosome 1 (49 cases) and gain of the X chromosome (44 cases). Clicking on 14q32 connects to the Map Viewer for this band, a region with 374 genes, and 8 CCAP BAC clones (Kirsch et al., 2000), which can be used for pinpointing the exact breakpoint in a particular specimen. This example of a particular search applied each parameter sequentially. It is also possible to search for multiple parameters at once by using the Advanced Search form by entering –1, 6p10, 14q32, 18q21, follicular lymphoma, and lymph nodes; this search brings up 20 cases.
TABLE 2.
Example of a Similarity Report: Search for 14q32 Results from first 500 Clones/Cells in Summary Report
| Total Clones/Cells 500 Karyotype 404 CGH 96 | |||||||||
| DIAGNOSIS | |||||||||
| Terms common to 100% of clones/cells | |||||||||
| None | |||||||||
| Terms common to 50%-99% of clones/cells | |||||||||
| None | |||||||||
| Terms common to <50% of clones/cells | |||||||||
| Follicular lymphoma | 109 | Renal cell carcinoma NOS C64.9 | 9 | ||||||
| Multiple myeloma | 52 | Carcinoma | 9 | ||||||
| Diffuse large B-cell lymphoma | 47 | Splenic marginal zone B-cell lymphoma | 8 | ||||||
| Burkitt lymphoma | 41 | Plasma cell leukemia | 8 | ||||||
| Mantle cell lymphoma | 25 | Adenocarcinoma, NOS | 8 | ||||||
| Acute lymphoblastic leukemia, NOS | 20 | Chronic lymphocytic leukemia | 7 | ||||||
| Papillary serous cystadenocarcinoma | 20 | T-prolymphocytic leukemia | 6 | ||||||
| Carcinoma, NOS | 18 | Squamous Carcinoma | 5 | ||||||
| Peripheral B-cell neoplasm | 17 | Leiomyoma | 4 | ||||||
| Acute lymphoblastic leukemia, FAB type L3 | 16 | Squamous cell carcinoma, NOS | 3 | ||||||
| BREAKPOINTS (Karyotype & CGH) | |||||||||
| Breakpoints common to 100% of clones/cells 14q32 | |||||||||
| Breakpoints common to 50%-99% of clones/cells | |||||||||
| None | |||||||||
| Breakpoints common to <50% of clones/cells | |||||||||
| 1p36 | 27 | 4q35 | 27 | 7q32 | 21 | 10q26 | 26 | 16q24 | 19 |
| 1q10 | 22 | 5p12 | 19 | 7q36 | 29 | 11q13 | 80 | 17q25 | 31 |
| 1q12 | 30 | 5q23 | 19 | 8p11.2 | 21 | 11q23 | 24 | 18q21 | 177 |
| 1q21 | 42 | 5q35 | 25 | 8q22 | 19 | 11q25 | 23 | 18q23 | 33 |
| 1q32 | 28 | 6p10 | 18 | 8q24 | 119 | 13q11 | 21 | 19q13 | 29 |
| 1q44 | 22 | 6q15 | 22 | 8q24.3 | 26 | 13q12 | 30 | 20q13.3 | 21 |
| 3p12 | 21 | 6q21 | 27 | 9p13 | 21 | 13q14 | 22 | 21q22 | 24 |
| 3q27 | 34 | 6q27 | 22 | 9q34 | 27 | 13q34 | 32 | 22q11 | 25 |
| 3q29 | 28 | 7q21 | 22 | 10q22 | 19 | 14q11.1 | 25 | 22q13 | 26 |
| 4p16 | 20 | 7q31 | 18 | 10q24 | 20 | 14q11 | 22 | Xq28 | 25 |
| Other categories in Similarity Report (data not shown): | |||||||||
| |||||||||
Note: This page is shown as it appears on the web site; underlined numbers of cases link to those particular cases, whereas underlined chromosome bands link to Map Viewer.
DISCUSSION
Many of the more than 500 molecular biology online databases (Galperin, 2004) were created and are maintained by the NCBI (Wheeler et al., 2004). The NCBI was established in 1988 to develop information systems for molecular biology, and the SKY/M-FISH & CGH Database and the Entrez Cancer Chromosomes database are part of this network. Cancer Chromosomes is part of NCBI’s Entrez integrated database retrieval system, which provides extensive links to related information.
SKY/M-FISH and CGH have markedly improved the ability to delineate chromosomal abnormalities (Kallioniemi et al., 1992; Schröck et al., 1996; Speicher et al., 1996). The SKY/M-FISH & CGH Database makes it possible to present the graphic depiction of all the abnormalities for all cases in any given study, thus enhancing the visual interpretation of the results; this is especially useful when the karyotypic and CGH profile results from the same case are analyzed. Comparing the cytogenetic profiles within and among tumors makes it possible to narrow down the genomic regions of relevance to tumor formation, and then to integrate the regions with the sequence databases.
The construction of the karyotype parser was complicated by the (linguistically unusual) conventions of the standard cytogenetic nomenclature (ISCN, 1995). The surface structure syntax, that is, how to write syntactically correct short forms, is well defined in the ISCN, and the parser program uses this information. The parser is able to transform both short-form and detailed-system (commonly referred to as the long form) karyotypes; the short form expresses an abnormal chromosome as a sequence of operators, if performed in sequence on a normal chromosome, will yield the abnormal chromosome, whereas the long form expresses an abnormal chromosome as an ordered list of named segments. The interpretation of short-form expressions is defined primarily through examples, but the ISCN does not have enough examples to cover the myriad rearrangements that occur in complex karyotypes. This lack of semantic interpretation contrasts with the practice for other formal languages. In most cases, the short-form karyotype appears to be unambiguous; however, it can be a challenge to use and interpret, and the system is error prone because of many complex rules; for those who are not cytogeneticists, it can also be difficult to understand. Another problem is that the short-form karyotype is what a computational linguist would call “deeply embedded”; this means that when reading or computationally processing a short-form karyotype, the meaning of a particular notation often depends, in a complicated way, on what has been seen or processed previously in that karyotype, and the extent of semantic processing may overtax short term memory. For very complex rearrangements, such as those revealed by SKY/M-FISH, where marker chromosomes contain material originating from many different chromosomes, the written karyotype in the long form is actually shorter than the short form and is easier to read. A new edition of the ISCN, now in progress, will address many of these issues, including how to report the chromosomal origin of hsrs and dmins as revealed by FISH, SKY/M-FISH, and CGH techniques.
By combining the results of clinical studies from the Mitelman Database with the clinical and research cases presented in the SKY/M-FISH & CGH Database, including cell line and mouse studies, the Cancer Chromosomes database makes it possible to perform an integrated search of a wide array of human and mouse cytogenetic data, and it provides, the first systematic way of placing chromosome aberrations on the sequence maps. It is a powerful search engine with potential application to the diagnosis and treatment of disease. The similarity tool retrieves commonalities among cases, data that are critical to the search for new genes such as oncogenes and tumor-suppressor genes, offering insights into the causes and consequences of genetic alterations in cancer.
Because the model used for CGH data is a subset of the karyotype data, it is now possible to examine directly the similarities between CGH results and the karyotypes presented in the Mitelman and SKY/M-FISH & CGH databases. All chromosomal bands, including breakpoints, are directly linked to the Map Viewer database, providing integration of cytogenetic data with map and sequence data from a variety of sources (Dombrowski and Maglott, 2002). Included within Map Viewer are the mapping positions of the high-resolution FISH-mapped BAC clones that were included in the CCAP initiative (Kirsch et al., 2000), which systematically integrated the cytogenetic and physical maps of the human genome. The ability to localize a particular breakpoint by use of the Cancer Chromosomes similarity tool can therefore identify specific BAC clones that can be used to pinpoint genes involved in particular cancers or in specific stages of tumor development and progression.
The refinement of these databases is not complete, and improvements in both presentation and function continue on a regular basis; new tools are added as they are developed and tested for reliability. Future plans include the integration of CGH array data, which currently can be submitted to the NCBI’s Gene Expression Omnibus database and to a gene expression and hybridization repository (Edgar and Lash, 2002). The ability to integrate cytogenetic data seamlessly with sequence will further the development of molecular tools to elucidate and comprehend the genetic instability of cancer.
Acknowledgments
The authors would like to extend their most sincere thanks to: Evelin Schröck, Joel Plotkin, and Carolyn Shenmen for their contributions to the initial development of the SKY/M-FISH & CGH Database; Jim Ostell for continuous support of the project; Felix Mitelman for allowing us to integrate his databases into Cancer Chromosomes; Leonid Khotomliansky, Susan Greenhut, and members of the Ried laboratory for database development and testing; the entire NCBI Entrez team, especially Pramod Paranthaman, Anton Golikov, Vladimir Sirotinin, and Grisha Starchenko; Aaron Ucko and Denis Vakatov from the C++ Team; database administrators Anthony Stearman and Slava Khotomlianski; Vyacheslav Chetvernin for help and discussions regarding the chromosome display libraries; Won Kim and John Wilbur for help with integrating and extending the document similarity algorithm; Sherri De Coronado for integrating ICD-O-3 into the NCI Metathesaurus; James Kriebel, Todd Groesbeck, and Buddy Chen for graphic design; and everyone who has submitted data to the SKY/M-FISH & CGH Database.
References
- Baudis M, Cleary ML. Progenetix. net: an online repository for molecular cytogenetic aberration data. Bioinformatics. 2001;17:1228–1229. doi: 10.1093/bioinformatics/17.12.1228. [DOI] [PubMed] [Google Scholar]
- Committee on Standardized Genetic Nomenclature for Mice, chairperson: Davisson MT. 1996. Rules for nomenclature of chromosome anomalies. In: Lyon MF, Rastan S, and Brown SDM, editors. Genetic Variants and Strains of the Laboratory Mouse, Third Edition, Vol 2, Oxford: Oxford University Press, p. 1443–1445.
- Dombrowski SM, Maglott D. Using the Map Viewer to Explore Genomes. 2002. In: McEntyre J, editor. The NCBI Handbook [Internet]. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information. Available from: http://ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
- Edgar R, Lash A. 2002. The Gene Expression Omnibus (GEO): A Gene Expression and Hybridization Repository. In: McEntyre J, editor. The NCBI Handbook [Internet]. Bethesda (MD): National Library of medicine (US), National Center for Biotechnology Information. Available from: http://ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
- Fritz A, Percy C, Jack A, Shanmugaratnam K, Sobin L, Parkin DM, Whelan S, editors. 2000. International Classification of Diseases for Oncology. Third Ed. Geneva: World Health Organization. 240 p.
- Galperin MY. The molecular biology database collection. Nucleic Acids Res. 2004 update;32:D3–D22. doi: 10.1093/nar/gkh143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huret J-L, Dessen P, Le Minor S, Bernheim A. The “Atlas of Genetics and Cytogenetics in Oncology and Haematology” on the Internet and a review of infant leukemias. Cancer Genet Cytogenet. 2000;120:155–159. doi: 10.1016/s0165-4608(99)00250-2. [DOI] [PubMed] [Google Scholar]
- ISCN (1995): An International System for Human Cytogenetic Nomenclature. 1995. Mitelman F, editor. Basel: S. Karger, 114 p.
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Kirsch IR, Green ED, Yonescu R, Strausberg RL, Carter NP, Bentley D, Leversha MA, Dunham I, Braden VV, Hilgenfeld E, Schuler DG, Lash AE, Shen GL, Martelli M, Kuehl WM, Klausner RD, Ried T. A systematic, high-resolution linkage of the cytogenetic and physical maps of the human genome: The Cancer chromosome aberration project (Ccap) Nature Genet. 2000;24:339–340. doi: 10.1038/74149. [DOI] [PubMed] [Google Scholar]
- Knutsen T, Gobu V, Knaus R, Ried T, Sirotkin K. 2002. The SKY/CGH database for spectral karyotyping and comparative genomic hybridization data. In: McEntyre J, editor. The NCBI Handbook [Internet]. Bethesda (MD): National Library of medicine (US), National Center for Biotechnology Information. Available from: http://ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
- Liyanage M, Coleman A, du Manoir S, Veldman T, McCormack S, Dickson RB, Barlow C, Wynshaw-Boris A, Janz S, Wienberg J, Ferguson-Smith MA, Schröck E, Ried T. Multicolour spectral karyotyping of mouse chromosomes. Nature Genet. 1996;14:312–315. doi: 10.1038/ng1196-312. [DOI] [PubMed] [Google Scholar]
- Mitelman Database of Chromosome Aberrations in Cancer (2003). Mitelman F, Johansson B and Mertens F, editors. http://cgap.nci.nih.gov/Chromosomes/Mitelman
- Ostell J. 2002. The Entrez search and retrieval system. In: McEntyre J, editor. The NCBI Handbook [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available from: http://ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI-60 Drug-Screening Panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- Speicher MR, Gwyn Ballard S, Ward DC. Karyotying human chromosomes by combinatorial multifluor FISH. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Suzek TO, Tatusova TA, Wagner L, Rapp Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]



