Abstract
Background
Colorectal cancer (CRC) is the third most prevalent cancer worldwide. Despite substantial advancements in CRC therapy in recent years, ionizing radiation (IR) continues to be the predominant treatment for colon malignances. However, it still lacks the precision required for excellent therapeutic outcomes, ultimately resulting in tumor radioresistance. This study seeks to explore the potential of atypical PARPs including PARP9, PARP12, PARP13 and PARP14 as innovative radiosensitizing targets for CRC.
Methods
We utilized CRISPR/Cas9-mediated gene editing to knockout the PARP9, PARP12, PARP13 and PARP14 in HT29 and DLD1 cells. The cells were exposed to either a single dose of 6–10 Gy or to fractionated dose of 5 × 2 Gy X-ray radiation cultivating cells in 2D, laminin-rich ECM 3D and multicellular spheroid models. The transcriptomes of nonirradiated and irradiated cells were analyzed using microarrays. Gene set enrichment analysis was conducted to determine the pathways in which PARP13 is engaged. Cell viability was assessed using a clonogenic assay. Gene expression levels in cells and patient samples were quantified using RT-qPCR.
Results
The expression of PARP9, PARP12, PARP13 and PARP14 was particularly elevated in irradiated colorectal cancer HT29 cells in a microenvironment-dependent manner. PARP13 deficiency significantly enhanced the sensitivity of HT29 cells to both single-dose and multifractionated irradiation regimens, resulting in reduced colony formation and spheroidal integrity. Microarray analysis indicated that PARP13 may modulate the expression genes associated with immune response signaling pathways, including members of PARP family. Furthermore, PARP13 loss in HT29 cells markedly impaired the expression of immune response related genes following multifractionated ionizing irradiation. Finally, chemoradiotherapy significantly elevated the expression of PARP9, PARP12, PARP13 and PARP14 in rectal tumors, while having no effect on adjacent normal colon tissues. Elevated pre-treatment PARP9 expression levels and a blunted post-treatment increase in PARP9 and PARP14 expression predicted poor overall survival in rectal cancer patients, while PARP13 emerged as the most significant discriminator between tumor and healthy tissue.
Conclusions
Collectively, the PARP9/13/14 axis is implicated in the response of CRC to radiation treatment in both preclinical and clinical settings, demonstrating the atypical members of the PARP family as attractive targets for neoadjuvant radiotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13046-025-03439-y.
Keywords: 3D cell culture, PARP9, PARP13, PARP14, Interferon stimulated genes, Radiotherapy, Radiosensitizers
Background
Colorectal cancer (CRC), a prevalent malignancy of the colon and rectum, ranks as the third most common cancer globally and is the second leading cause of cancer-related deaths [1]. However, colorectal cancer is often diagnosed as Stage II or above when treatment often necessitates a combination of chemoradiotherapy and surgical treatment [2]. While CRC therapy has made significant progress in recent years, ionizing radiation (IR) remains a current standard therapy for colon malignancies [3]. IR induces DNA damage in tumor cells, including abasic lesions, deoxyribose ring opening, single and double-stranded breaks (SSBs and DSBs), ultimately leading to cell death [4]. Conversely, tumor cell resistance to IR hinges on their ability to trigger a cascade of cellular responses as a part of DNA damage response (DDR) and a successful DNA damage repair [5]. Advancements in radiotherapy equipment and treatment planning enable the shaping of radiation dose to target tumors while minimizing exposure to surrounding healthy tissues. Despite its potency, radiotherapy can lack the precision required for optimal therapeutic outcomes, ultimately leading to tumor radioresistance. To address this challenge, the discovery of novel biological therapy targets that could enhance damage to cancer cells while significantly sparing normal tissue cells remains increasingly important.
Among the key players in the DDR networks are members of the 17 PARP (poly-ADP-ribose polymerase) protein family. These proteins typically exhibit ADP-ribosylation activity, transferring nicotinamide adenine dinucleotide (NAD+) to target molecules. Beyond their involvement in various biological processes - such as transcription control, cell death, and inflammation [6]– some PARPs specifically contribute to attracting repair proteins to sites of DNA damage [7]. PARP1, the most well-studied member, acts as a DNA damage sensor and signal transducer synthesizing poly-ADP-ribose chains that recruit DNA repair factors and remodel chromatin structure around damaged DNA [8]. Consequently, targeting PARP activity has gained prominence in cancer therapy, as the first synthetic lethal targeted therapy. Successful inhibition of PARP1 activity leads to unrepaired SSB accumulation, followed by DSB generation, therefore combining PARP inhibitors with treatment targeting tumors possessing defective DDR systems hold the potential for creating effective combination therapy strategies [9]. Notably, recent studies have highlighted that PARP inhibitors could sensitize tumors to radiotherapy [10–12].
In addition to well-characterized PARPs, other members of this family hold promise for cancer therapeutic applications as they also take part in DNA damage response [13, 14]. For instance, PARP13 scaffolds PARP1-HSF1 complex at DNA break sites to recruit DNA repair machinery [13], while PARP14 is a critical factor in stabilizing stalled replication forks, particularly in cells with BRCA1/2 deficiencies [14]. Moreover, lack of PARP14 impairs homologous recombination-mediated repair of double-strand breaks, resulting in increased sensitivity to DNA damaging agents [15, 16]. PARP9, recruited to DNA damage sites, forms a complex with DTX3L, contributing to genome integrity through targeted ubiquitination of p53 [17]. Meanwhile, the precise role of PARP12 in DNA damage repair remains to be fully elucidated, its interaction with PARP1 suggests a potential involvement in the process [18, 19]. Therefore, the initial evidence on lesser-explored PARP family members indicates that these proteins may serve as promising targets for radiosensitization and could open a potential window for the application of adjuvant therapy following radiotherapy.
While two-dimensional (2D) monolayer cell culture models have been invaluable in cancer research, they often fall short in capturing the complexities of the tumor microenvironment. Three-dimensional (3D) cell cultures, however, offer a more accurate representation of in vivo cancer tissue by mimicking the spatial architecture, nutrient gradients, and cell-cell interactions observed with solid tumors [20]. In addition, the tumor microenvironment modulates cancer immunity features, influences cell shape, proliferation, gene expression and response to therapy [21]. As a result, to enhance comprehension of the complexities of tumor response to treatment and to propose innovative paths for cancer therapy development, the utilization of 3D cell culture models is indispensable.
We previously discovered that the multifractionated dose IR upregulated the expression of unconventional PARP family members including PARP9, PARP12, PARP13 and PARP14 in colorectal cancer cells grown in laminin-rich ECM 3D cell culture [22]. Currently, there is no consolidated data on how PARP9, PARP12, PARP13 and PARP14 affect the response to ionizing radiation in cancer cells. Consequently, this study exploited comprehensive transcriptome analysis to evaluate the potential application of these atypical PARPs as radiosensitizers for colorectal cancer cells in 3D cell culture models. Finally, we conclude that selected PARPs are involved in the response of CRC to radiotherapy, indicating that these proteins are promising targets for the improvement of future treatment options.
Methods
Cell line maintenance
Human normal colon tissue CRL1790 and colorectal carcinoma DLD1, HT29, HCT116 and SW48 cell lines were obtained from the American Type Culture Collection (Rockville, Maryland, USA). Cells were maintained in RPMI-1640 (DLD1, HCT116) or DMEM (CRL1790, HT29, SW48) cell culture media (Thermo Scientific, USA) supplemented with 10% fetal bovine serum (Biochrom, Germany), 2 mM glutamine (Thermo Scientific), 100 U/ml penicillin (Carl Roth, Germany) and 0,1 mg/ml streptomycin (Carl Roth, Germany). Cells were incubated at 37 °C in a humidified atmosphere containing 5% CO2.
Cell culture models
For the 2D cell culture, CRL1790, DLD1, HT29, HCT116 and SW48 cells were plated in T25 flasks (30000 cells/cm2). For lr-ECM 2D cell culture model, 25,000 CRL1790 cells were embedded in 3.2% lr-ECM protein mixture Geltrex (Thermo Scientific) in DMEM mixture and seeded into 24 well plates pre-coated with 10% Geltrex in DMEM mixture. For lr-ECM 3D cell culture, 75,000 of DLD1 and HT29 cells were embedded in 3.2% Geltrex in culture media in 24 well plates as described previously [23]. Multicellular spheroids (MCS) were formed using liquid-overlay technique as described previously [24]. Briefly, cells were suspended in 200 µl cell culture medium and plated in each well of 96 U-bottom well plates and centrifuged at 1000 g for 10 min. To avoid cell attachment to the well bottom, each well was pre-coated with 1% agarose in sterile water. Cell number established empirically for each cell line to form 400 ± 20 μm size spheroids 2 days after plating. Cells were photographed every two days with inverted optical microscope Eclipse TS100 using digital camera DS-Fi2 (Nikon, Japan).
Patient samples
The institutional review board approved the study. Patients diagnosed with rectal cancer received neoadjuvant long-course chemoradiotherapy which included 25–28 fractions of irradiation (total dose of 45–51 Gy) and fluorouracil-based treatment during a 5-week period. Tumor and adjacent normal rectal tissue samples were obtained by biopsy and tumor resection surgery 8 or 12 weeks after the neoadjuvant treatment and stored at − 80 °C in RNAlater (Thermo Scientific) until needed. The sample cohort contained four groups and included rectal tumor (n = 67), and adjacent normal (n = 31) tissue samples collected from patients before long course neoadjuvant treatment (biopsies) and samples collected from the same patients after the therapy (during the surgery). Patient demographic and clinical characteristics are summarized in Table 1.
Table 1.
Demographic and clinicopathological characteristics of rectal cancer patients
| Factor | Total | % | |
|---|---|---|---|
| Age (median, range) | 68 (41–87) | ||
| Sex | Male | 36 | 53.7 |
| Female | 31 | 46.3 | |
| Stage | 1 | 2 | 3 |
| 2 | 5 | 7.5 | |
| 3 | 59 | 88.1 | |
| 4 | 1 | 1.5 | |
| Pretreatment staging | |||
| cT | 2 | 5 | 7.5 |
| 3 | 51 | 76.1 | |
| 4 | 11 | 16.4 | |
| cN | 0 | 4 | 6 |
| 1 | 16 | 23.9 | |
| 2 | 47 | 70.1 | |
| Pathological staging | |||
| ypT | 0 | 8 | 11.9 |
| 1 | 2 | 3 | |
| 2 | 17 | 25.4 | |
| 3 | 35 | 52.2 | |
| 4 | 5 | 7.5 | |
| ypN | 0 | 43 | 64.2 |
| 1 | 17 | 25.4 | |
| 2 | 7 | 10.4 | |
| Dworak Tumor Regression Grading | 1 | 19 | 28.4 |
| 2 | 31 | 46.3 | |
| 3 | 9 | 13.4 | |
| 4 | 8 | 11.9 | |
| Interval between neoadjuvant chemoradiation completion and surgery (weeks) | 8 | 30 | 44.8 |
| 12 | 37 | 55.2 | |
CRISPR/Cas9 editing and clone selection
sgRNAs were designed using Benchling CRISPR sgRNA Design tool [25]. sgRNAs sequences are shown in Additional Table 1. For transient expression of the CRISPR/Cas9 system, sgRNA nucleotides were cloned into pSpCas9(BB)-2 A-GFP (purchased from Addgene, #48138) at BbsI site as described previously [26]. Cell transfection was carried out using Turbofect (Thermo Scientific) following the manufacturer’s protocol. Briefly, DLD1 and HT29 cells were plated into 6-well plates (1 × 106 cells/well) 24 h prior transfection. Next, a mixture containing 2 µg of pSpCas9(BB)-2 A-GFP plasmid constructs and 10 µL of Turbofect in final volume of 300 µl Opti-MEM (Thermo Scientific) growth medium was incubated at room temperature for 20 min and added to the cells. 48 h following cell transfection, GFP positive cells were analyzed and sorted using BD FACS AriaIII™ flow cytometer (BD Biosciences, USA). Single cell colonies were obtained by serial dilution of sorted cells plated in 96 well cell culture plates. Generated monoclonal cells were lysed using Phire Tissue Direct PCR Kit (Thermo Scientific) according to the manufacturer’s instructions. Briefly, cell pellets were lysed in 20 µl of lysis buffer including 0.5 µl DNA Release Additive at 98 °C for 5 min. Genomic PCR was carried out using 0.75 µl of cell lysates and the primers listed in Additional Table 2. PCR amplification was carried out in a SensoQuest Labcycler (Germany). Lastly, genome editing was verified performing T7 endonuclease assays according to manufacturer’s instructions (New England Biolabs). Successfully modified clones were selected for further experiments.
Immunoblotting
Total protein was extracted from cells 48 h after plating (1 × 106 cells per T25 flask). Cells were lysed in RIPA lysis buffer supplemented with 1x protease inhibitor (Thermo Scientific). Western blot analysis was performed by standard immunoblotting following SDS-polyacrylamide gel electrophoresis using 4% stacking gels and 8% separating gels. Protein extracts were analyzed with the following primary antibodies: PARP9 (Thermo Scientific, cat. no. 40-4400; 1:1000), PARP12 (Thermo Scientific, cat. no. PA5-51967; 1:1000), PARP13 (Thermo Scientific, cat. no. PA5-31650; 1:5000), PARP14 (Santa Cruz, cat. no. sc-377150; 1:1000), actin (Abcam, cat. no. ab8227; 1:10000). The following secondary antibodies were used at a 1:1000 dilution: Rabbit IgG HRP-conjugated (R&D Systems, cat. no. HAF008) and Mouse IgG HRP-conjugated (R&D Systems, cat. no. HAF018). Visualization of membranes was performed using Intas Chemocam Imager system (Royal Biotech, Germany).
Cell irradiation
Cells were irradiated at room temperature with 6MV X-rays using Varian clinical linear accelerator (Clinac 600 C/D; Varian Medical Systems Inc., Palo Alto, CA, USA). Two different irradiation regimes were used: a single dose of 6–10 Gy and a fractionated dose delivered daily in 5 fractions, 2 Gy per fraction (5 × 2 Gy). The 10 Gy single dose was administered only in the initial evaluation of PARP gene expression, whereas subsequent experiments facilitated the administration of a single 6 Gy dose, since its biologically effective dose (1 × 6 Gy BED = 18.0) is more comparable to 5 × 2 Gy regimen (BED = 16.7 Gy). The dose rate was ~ 3 Gy/min. In all experiments the same experimental design and separate controls of nonirradiated cells were used for all regimens.
Clonogenic survival assay
DLD1 and HT29 cells were plated in 6‑well plates for 2D (500 cells/well) or embedded in 96 well plates for 3D (500–1000 cells/well) 24 h prior to irradiation and treated with single dose of 6 Gy or fractionated dose of 2 Gy of ionizing radiation (IR) daily for 5 days. In total, 8 days after irradiation DLD1 and HT29 cell colonies (> 50 cells/colony) were fixed with methanol, stained with crystal violet and counted manually. Clonogenic survival was evaluated as described previously [22]. The mean cell survival fraction from 3 independent experiments was used to represent survival at each irradiation dose.
Spheroid growth assay
DLD1 and HT29 cell spheroids were plated 48 h prior to irradiation. Control and irradiated spheroids were cultivated on separate plates throughout the duration of experiments (overall 14 days including the plating of MCS). Every two days post irradiation, MCSs were imaged with an inverted optical microscope Eclipse TS100 using digital camera DS-Fi2 (Nikon, Japan). Spheroid size was evaluated using SpheroidSizer 1.0.
MCS cell survival assay
The viability of DLD1 and HT29 cell speroids was measured using a CellTiter-Glo 3D Cell Viability Assay kit (Promega, USA) following the manufacturer’s instructions. Briefly, 48 h after the final fraction of 5 × 2 Gy irradiation the individual spheroids were transferred into individual wells of a white opaque 96-well plate (Thermo Scientific). CellTiter-Glo 3D reagent was then added to each well and the intensity of chemiluminescence was measured 30 min later using the Variokan LUX multiplate reader (Thermo Scientific). In parallel, non-irradiated MCS were utilized as controls.
RNA extraction
Cells, cultured in 2D, lr-ECM 2D, lr-ECM 3D, or MCS conditions, were harvested after 2 or 6 days of growth, depending on the experimental design performed. In cases involving cell irradiation (single dose or multifractionated) RNA was extracted 4 h after the final dose delivery. Total RNA was isolated using Quick RNA MiniPrep Kit (Zymo Research) following manufacturer’s instructions. For rectal tissue samples, total RNA was isolated using MirVana miRNA isolation Kit (Thermo Scientific) as per manufacturer’s instructions. The quantity and quality of RNA were evaluated using Nanodrop 2000 (Thermo Scientific). Subsequent RNA integrity assessment, based on the evaluation of 28 S and 18 S ribosomal subunit peaks in electropherogram, was performed using Bioanalyzer 2100 (Agilent, USA).
Microarray analysis
cRNA sample preparation, labeling and hybridization was performed according to manufacturer’s instructions. Briefly, 100 ng of total RNA was used for cDNA synthesis and amplification using Low Input Quick Amp Gene Expression Labeling Kit (Agilent). Then 825 ng of cRNA labeled with Cy3/Cy5 dyes using Gene Expression Hybridization Kit (Agilent) were hybridized to Human 4 × 44k Oligonucleotide Microarrays (Agilent) using HS 400 hybridization station (Tecan, Switzerland). Microarray slides were scanned using SureScan Microarray scanner (Agilent). Microarray image analysis and data generated were further analyzed using ImaGene ver. 9.0 (BioDiscovery, USA) and GeneSpring GX v11.5 software (Agilent, USA). Loess normalization was performed to adjust microarray data for variation. Gene expression fold change above 1.5 (with p-value < 0.05) was defined as differentially expressed between two conditions. Microarray design and data are available at the GEO database (Accession No. GSE280331, GSE280332).
Microarray data enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of gene expression data was performed using the WEB‑based Gene SeT AnaLysis Toolkit, as described previously [27]. P‑values were calculated using the hypergeometric test and adjusted using the Benjamini and Hochberg procedure. Functional KEGG pathway categories associated with ≥ 5 genes were considered as significantly enriched (p < 0.05) in differentially expressed genes.
RT-qPCR
cDNA was synthesized using Revert Aid RT Kit (Thermo Scientific) according to manufacturer’s instructions. A total of 1 µg RNA was used for cDNA synthesis. Quantitative real-time PCR was performed using QuantStudio5 thermal cycler (Applied Biosystems, USA) and Maxima SYBR Green qPCR Master Mix (Thermo Scientific) according to manufacturer’s instructions. The relative changes in gene expression were calculated by ΔΔCt method using GAPDH as the housekeeping gene (for cell line experiments) or GAPDH, ACT and TBP geomean (for patients’ tissue samples) for sample normalization [28]. Primer sequences are shown in Additional Table 2.
Statistical analysis
The statistical analysis was performed using the data analysis software package SPSS 26.0 (IBM) and GraphPad v9.0 (GraphPad Software). The statistical significance of gene expression and surviving fraction differences in cells was calculated using an unpaired t-test. For differences of spheroid growth kinetics evaluation, two-way ANOVA was used. Each data point is displayed as the mean ± standard deviation of at least three independent biological experiments. Normal distribution of data was verified using Shapiro-Wilk test. Differences in PARP expression levels in rectal tissue samples were evaluated using paired t-test. Patients were separated into two groups based on gene mean expression (cut-off value). Samples with less than mean expression level were assigned to the low expression group; samples with above mean value were assigned to the high expression group. A two-sided Chi-square test or Fisher’s exact test were used to analyze the distribution of cases with low or high studied genes level in tumor samples according to the demographic and clinicopathological characteristics of the patients. The overall survival (OS) and progression-free survival (PFS) were evaluated by Kaplan-Meier analysis and log-rank test. Univariate and multivariate Cox regression analysis was performed to detect independent factors significantly determining OS or PFS. Bioinformatic analysis of TCGA data (expression and survival) was performed by GEPIA2 online software [29]. The observations with a p-value of less than 0.05 were considered significant.
Results
Ionized radiation affects PARP gene expression in colorectal cancer cells in a microenvironment dependent manner
Guided by genome-wide transcriptomics data [22] in colorectal cancer cells exposed to single (2–10 Gy) or multifractionated of a 2 Gy dose of X-ray ionizing irradiation every 24 h for 5 days (Fig. 1A), we selected four PARP genes including PARP9, PARP12, PARP13 and PARP14 and implemented a more detail analysis of selected gene expression levels in non-irradiated cancer and non-cancerous cell lines and cells subjected to multifractionated 5 × 2 Gy irradiation (Fig. 1B). The experiments were performed using three different cell plating conditions: DLD1, HT29, HCT116 and SW48 cells were grown as plastic-attached monolayer (2D), scaffold-attached cells in laminin-rich extracellular matrix (lr-ECM 3D) or scaffold-free multicellular spheroids (MCS) (Fig. 1B, Additional Fig. 1A). For normal colon CRL1790 cells, we selected a monolayer model system with a laminin-rich ECM pre-coated surface (Additional Fig. 1B).
Fig. 1.
Ionizing radiation induces PARP gene expression in colorectal cancer cells in a microenvironment dependent manner. (A) Heatmap analysis depicts the expression pattern of PARP family genes in laminin-enriched 3D cell cultures of DLD1 and HT29 cells following multifractionated (5 × 2 Gy) dose ionizing radiation treatment. Unirradiated cells were used as a control. The microarray dataset originates from a previous study (GSE75551). (B) Multifractionated irradiation experimental design. 24 h after cell plating, cells were irradiated with a single dose of X-ray (2 Gy) every 24 h for 5 days, resulting in a total dose of 10 Gy. RNA for gene expression analysis was extracted 4 h after cell irradiation. Below are schematics and representative phase-contrast images of CRC cells grown under three different plating conditions: monolayer culture (2D), three-dimensional laminin rich-extracellular matrix culture (lr-ECM 3D), and multicellular spheroid (MCS) system. Scale bars indicate 200 μm. (C) Expression of PARP9,12,13,14 genes was examined using RT-qPCR in colorectal cancer cells (DLD1, HT29) cultivated under 2D or both 3D cell culture conditions after exposure to multifractionated irradiation (5 × 2 Gy). (D) Similarly, the expression of PARP9,12,13,14 was investigated in normal colon cells (CRL1790) grown in a monolayer or lr-ECM coated 2D culture, following the same regimen of irradiation (5 × 2 Gy). Results show means with error bars representing standard deviation (n = 3, *p < 0.05, Student’s t-test)
Quantitative PCR analysis revealed that multifractionated irradiation significantly upregulated PARP9, PARP12, PARP13 and PARP14 expression in HT29 cells cultured in both lr-ECM 3D and MCS (FC > 1.5, p < 0.05) (Fig. 1C) whereas no significant change in expression was observed in cells grown as a monolayer compared to non-irradiated cells. In contrast, DLD1, HCT116 and SW48 cells did not show such PARP expression response to multifractionated irradiation (Additional Fig. 2). Interestingly, irradiated normal colon cells exhibited only a modest effect on PARP expression when cultured in 2D but not in lr-ECM (Fig. 1D). Altogether, these preliminary findings suggest that the expression of PARP9, PARP12, PARP13 and PARP14 in irradiated cells is reliant on 3-dimensional cellular microenvironment or cell type, and the PARP expression pattern may be specific to cancer cells.
Fig. 2.
PARPs regulate colorectal cancer cell radiosensitivity in a microenvironment-dependent manner. (A) DLD1 and HT29 cell sublines with indicated PARP knockouts were generated using CRISPR/Cas9 genome editing. Single dose and multifractionated irradiation experimental design for colony formation assay (1) and MCS growth assay (2). (B) Clonogenic survival of CRC PARP knockout cells after irradiation with a single dose (6 Gy) or multifractionated (5 × 2 Gy) regimens. Wild type cells were used as the control. Results show means with error bars representing standard deviation (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t-test). (C) Morphology of CRC PARP knockout cells spheroids, (D) their growth kinetics and viability following multifractionated irradiation treatment. Representative images show the spheroids grown at the optimal seeding densities between day 0 and 6 of irradiation treatment. Scale bars indicate 200 μm. The growth kinetics are represented by spheroid diameter on the 6th day of irradiation treatment; same time point for MCS viability. Unirradiated spheroids were used as the control. Results show means with error bars representing standard deviation (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, during statistical evaluation, two-way ANOVA for MCS diameter, Student’s t-test for MCS viability)
PARP13 is the most prominent modulator of CRC cell sensitivity to multifractionated irradiation
To investigate the role of PARP genes in colorectal cancer cell response to ionizing irradiation, we generated PARP12, PARP13, and PARP14 knockout (KO) sublines of DLD1 and HT29 cells using CRISPR/Cas9 gene editing (Additional Fig. 3). The produced PARP KO subclones, along with the WT control lines, were evaluated for cell viability in 2D and lr-ECM 3D cell culture settings after the irradiation with a single (6 Gy) or multifractionated (5 × 2 Gy) dose as shown in Fig. 2A. Colony formation analysis (Fig. 2B, Additional Fig. 4) revealed that DLD1 PARP KO cells were as radiosensitive as wild type cells when treated with a single dose of ionizing radiation in both cell culture conditions, whereas multidose exposure significantly impaired the survival of PARP12(−/−), PARP14(−/−) cells grown in monolayer, but not in lr-ECM 3D. In contrast, HT29 cells exhibited a distinct profile of PARP-dependency on the response to ionizing irradiation. While only PARP13 KO dramatically increased the susceptibility of HT29 cells to a single-dose irradiation in 2D, monolayer HT29 PARP12(−/−), PARP13(−/−) and PARP14(−/−) sublines exhibited significantly reduced survival upon exposure to 5 × 2 Gy. Notably, PARP13 KO had the most dramatic effect, reducing the surviving cell fraction down to 7.4%. Remarkably, a significant level of radiosensitization was retained in HT29 PARP12(−/−) and PARP13(−/−) within lr-ECM 3D cultures when exposed to multifractionated irradiation, exposing PARP13 KO cells as more susceptible to radiation than PARP12 KO cells as well. Overall, these results indicated that PARP13 KO had the most pronounced effect on HT29 cell radiosensitivity, significantly reducing colony formation in both 2D and lr-ECM 3D growth conditions (p < 0.001 and p < 0.01, respectively) when using a clinically relevant radiotherapy regimen.
Fig. 4.
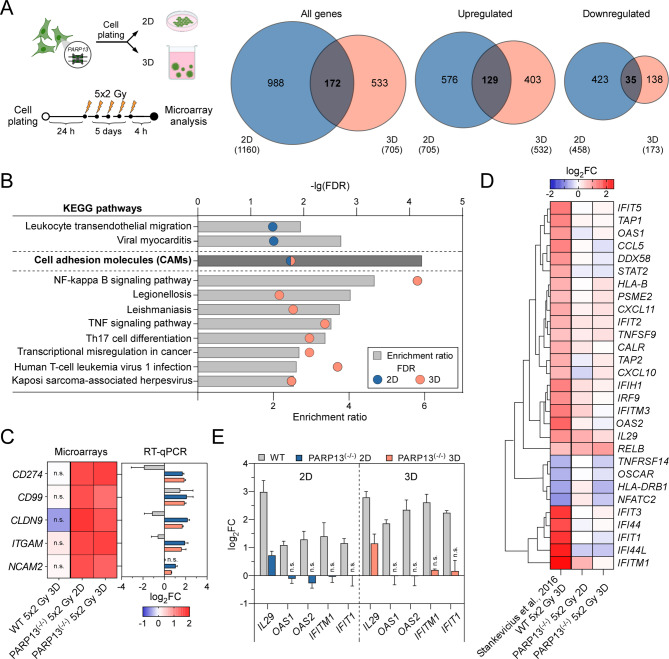
PARP13 regulates immune gene expression in ionizing radiation treated CRC cells. (A) Experimental design of microarray analysis for multifractionated irradiation treated HT29 PARP13 knockouts in both 2D and lr-ECM enriched 3D conditions. Cells were irradiated with a single dose of X-ray (2 Gy) every 24 h for 5 days, resulting in a total dose of 10 Gy. Total RNA was extracted 4 h post cellular irradiation. Venn diagrams illustrate differentially expressed genes (DEGs) in 2D (blue) and lr-ECM 3D (pink) conditions from global analysis in irradiated HT29 PARP13 knockouts relative to non-treated PARP13 knockout cells, determined using Agilent array analysis of total mRNA (n = 3 independent experiments). (B) Significantly enriched KEGG pathways in irradiated PARP13 knockout cells grown under 2D and lr-ECM 3D conditions. The upper X-axis represents -lgFDR values, the lower X-axis indicates the enrichment ratio, and the Y-axis shows the enriched KEGG pathways. (C) Microarray data validation using RT-qPCR analysis. Results are presented as means with error bars representing standard deviation (n = 3, Student’s t-test, n.s.– not significant). (D) Clustering heatmap of immune response related gene expression data in irradiated HT29 wild type (GSE75551) and PARP13 knockout cells under 2D or lr-ECM enriched 3D conditions. (E) Confirmation of differential expression of selected four immune response-related genes in HT29 wild type and PARP13 knockout irradiated cells using RT-qPCR. Results demonstrate means (n = 3, Student’s t-test, n.s.– not significant)
To further validate the impact of PARPs on colorectal cancer cell susceptibility to ionizing radiation, we irradiated CRC multicellular spheroids with 5 × 2 Gy and monitored the MCS morphological integrity and growth rate dynamics over a two-week period (Fig. 2A). Surprisingly, the initial spheroid growth analysis revealed that PARP13 deficiency affects spheroid size even before irradiation, as HT29 PARP13 KO spheroids were the smallest in diameter among all PARP KO variants compared to wild type cells (Additional Fig. 5). On the other hand, irradiation did not affect DLD1 WT and PARP13(−/−) spheroid structural integrity, while PARP12(−/−) and PARP14(−/−) spheroids started to disintegrate 6 days after treatment initiation. In contrast, all HT29 PARP KO spheroids exhibited visible disintegration at the edge level (Fig. 2C, Additional Fig. 7A). As expected, HT29 PARP13(−/−) spheroids were particularly susceptible to disintegration, suggesting a role for PARP13 in maintaining spheroidal structure via cellular adhesion or cellular survival. Moreover, HT29 PARP12(−/−) and PARP13(−/−) spheroids experienced a considerable reduction in size following irradiation (Fig. 2D and Additional Fig. 6), supporting the impact of PARPs on colorectal cancer cell sensitivity to IR in the microenvironment dependent manner. Supporting these observations, spheroid viability assays confirmed that HT29 PARP13 cells exhibited the greatest sensitivity to fractionated dose irradiation, while knockouts of other PARP genes did not significantly alter radiation sensitivity in either HT29 or DLD1 cells (Fig. 2D, Additional Fig. 7B and C). These results, validated by both spheroid size and viability assays, underscore the role of PARP13 in modulating cellular response to ionizing radiation.
Fig. 5.
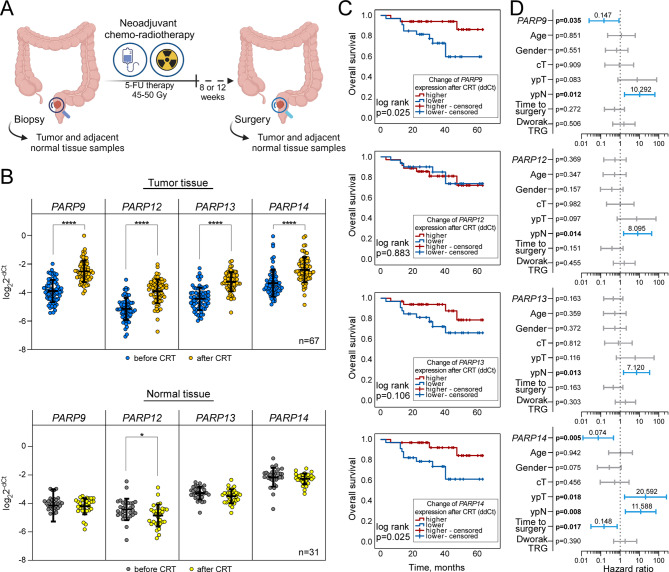
Association of PARP9,12,13,14 gene expression with rectal cancer clinical outcome following chemoradiotherapy (CRT) treatment. (A) Schematic overview of sample collection from rectal cancer patients. Tumor and adjacent normal tissue samples were obtained through biopsies patients with suspected rectal cancer. Following confirmation of diagnosis, patients underwent neoadjuvant chemoradiotherapy involving 5-fluorouracil before surgical resection of tumor. During the tumor resection surgery, additional samples of tumor and adjacent normal tissue were collected from each patient for further analysis. (B) qRT-PCR analysis of relative PARP gene expression levels before and after CRT in tumor and normal tissue sample groups. The cycle threshold (Ct) values of target genes were normalized to GAPDH, ACT and TBP levels. Lines within boxes indicate relative gene expression mean values, while whiskers denote standard deviation of the relative gene expression values (tumor n = 67, normal n = 31, Student’s t test, *p < 0.05 and ****p < 0.0001). (C) Kaplan-Meier survival curves demonstrating the association between changes in PARP expression after CRT and overall survival (OS) in rectal cancer tissue samples (n = 67). Patients were stratified into high and low change of expression groups according to the mean value. Curves were compared using the log-rank test, p values shown. (D) Prognostic performance of PARP genes expression changes and clinicopathologic features by multivariate Cox regression analysis. Forest plot illustrates the hazard ratio (vertical bar and number above it), and 95% confidence intervals (whiskers) associated with predictors for rectal cancer patients’ OS in tumor samples (n = 67). Significant predictors are highlighted in blue, with displayed p values
Additionally, a parallel survival analysis showed that reduced levels of PARP9 in HT29 cells (Additional Fig. 8A and B) were associated with reduced viability after a single 6 Gy irradiation, while no other changes in radiosensitivity were observed in cells grown in 3D cell models compared to WT cells (Additional Fig. 8C and D).
Overall, our observations underscored the radiosensitizing effect of selected PARPs which was cancer cell type and cellular microenvironment dependent proposing PARP13 as an essential gene for maintaining HT29 cell survival under the stress of radiation exposure. PARP13(−/−) cells exhibited increased sensitivity to both single-dose and multifractionated ionizing radiation regimens, leading to an impaired survival and spheroidal integrity.
PARP13 regulates the expression of genes associated with immune response in CRC cells
Having demonstrated that the loss of PARP13 is associated with increased CRC cell sensitivity to multifractionated irradiation in a microenvironment dependent manner, we went on to the level of whole genome transcriptome to compare the relative abundance of transcripts in HT29 PARP13(−/−) cells to wild type cells cultured in 2D and lr-ECM 3D model systems (Fig. 3A). Microarray analysis revealed significant changes in gene expression, with 667 and 536 differentially expressed genes (DEGs) in 2D and lr-ECM 3D, respectively, exposing a lesser deregulation of gene expression in 3D cultures. Conversely, a higher number of genes were downregulated in 3D as well. Further analysis of overlapping 243 DEGs between cells cultured in 2D and 3D showed a slightly higher number of upregulated genes suggesting PARP13-related relaxation of specific transcripts.
Fig. 3.
PARP13 regulates immune gene expression in CRC cells. (A) Experimental design of microarray analysis. Total RNA was extracted 48 h after plating HT29 wild type and PARP13 knockout cells in 2D (blue) or lr-ECM 3D (yellow) culture conditions. Venn diagrams illustrate differentially expressed genes (DEGs) in HT29 PARP13 knockout cells compared to wild type cells, determined using Agilent array analysis of total mRNA (n = 3 independent experiments). (B) KEGG pathways significantly enriched in HT29 PARP13 knockout cells grown under 2D and lr-ECM 3D conditions. Shared pathways commonly enriched in both cell culture conditions are highlighted in bold. The upper X-axis represents -lgFDR values, the lower X-axis indicates the number of genes in each pathway. The Y-axis shows the enriched KEGG pathways. (C) KEGG pathway enrichment analysis of overlapped genes between 2D and lr-ECM 3D systems, encompassing both upregulated and downregulated genes. The Y-axis represents significantly enriched KEGG pathways, the X-axis displays the enrichment ratio, and the color of the dots signifies -lgFDR values. The size of the dots corresponds to the number of genes enriched in the respective KEGG pathways. (D) Comparative Venn’s diagram analysis showing amount of DEGs in comparison to an independent PARP13 knockout experiment in HeLa cell line (GSE56667). The box emphasizes genes commonly expressed among all three groups. (E) Microarray data validation using RT-qPCR analysis. Bars indicate fold change of representative upregulated genes belonging to three distinct groups: PARP family, apoptosis, and immune response in HT29 PARP13 knockout cells compared to wild type cells. The results display means with error bars representing standard deviation (n = 3, p < 0.05, Student’s t-test)
The latter KEGG analysis showed that overlapping genes were primarily involved in multiple pathways associated with immune response (Fig. 3B, Additional Tables 3 and 4). Notably, these pathways were more significantly enriched in DEGs when cells were cultivated in 3D culture (Fig. 3C, Additional Table 5), suggesting that PARP13 could regulate cancer cell immune response signaling in a microenvironment dependent manner.
To further assess the specificity of PARP13 related gene deregulation, we also compared our DEG list with gene expression data set resulted from PARP13 silencing experiments in HeLa cells [30] (Fig. 3D). Although we identified a set of only 25 DEGs common among three compared gene sets, the profile was strongly associated with immune pathways indicating a significant role for PARP13 in the regulation of cellular immune responses cancer cell lines of different origins.
To verify the microarray data, we selected genes associated with PARP family (PARP4, PARP9, PARP10, PARP14), apoptosis (TNFSF10, TNFRSF10B, STAT1, STAT2, IRF9), and immune response pathways (IFIT1, IFIT2, IFIT3, OASL, ISG15). qRT-PCR results largely confirmed the microarray data trends observed in HT29 cells under both 2D and lr-ECM 3D growth conditions (Fig. 3E). Notably, PARP13 itself emerged as a regulator of other PARP family members expression levels, with the strongest effect on PARP9 and PARP14. Furthermore, HT29 PARP13(−/−) cells exhibited induced expression of known proapoptotic genes TNFSF10, TNFRSF10B and IRF9 proposing a pro-survival function for PARP13. Additionally, the highest upregulation was observed in immune response genes, further highlighting the potential involvement of PARP13 in immune regulation. While the microarray data showed no clear difference in gene expression levels between 2D and 3D cultures for the selected genes (Additional Fig. 9A), qRT-PCR validation revealed some discrepancies. Notably, the increase in OASL and ISG15 expression, genes related to the immune response, was lower in 3D compared to 2D. In contrast, PARP13 status did not correlate with expression of PARP family genes in DLD1 cells (Additional Fig. 9B), while the expression of apoptosis and immune pathways-related genes showed a much lower response compared to HT29 cells suggesting that PARP13 regulatory targets might be cell type dependent.
Taken together, these findings indicate that PARP13 status in CRC cells correlates with the expression regulation of genes associated with immune response signaling pathways, potentially impacting the PARP family regulation itself. Furthermore, PARP13’s role in the regulation of survival genes in CRC cells might be microenvironment dependent.
Loss of PARP13 impairs immune response in CRC cells triggered by multifractionated radiation
Given that PARP13 might impact CRC cell radiosensitivity in a microenvironment dependent manner, we sought to expose core molecular pathways influenced by PARP13-dependent regulation in irradiated HT29 PARP13(−/−) cells grown in both 2D and lr-ECM 3D cultures (Fig. 4A). The microarray analysis yielded a higher number of differentially expressed genes in 2D cultured cells (1160) compared to 3D (705), of which only 172 DEGs overlapped between both growth conditions, suggesting that cellular response is significantly microenvironment dependent. Notably, KEGG analysis revealed that DEGs resulting from both conditions were primarily enriched in cell adhesion molecules (CAMs) (Fig. 4B, Additional Tables 6 and 7). The following RT-qPCR analysis of selected five CAM genes (CD274, CD99, CLDN9, ITGAM, NCAM2) confirmed a potential role for CAMs in the response to irradiation with PARP13 deficiency (Fig. 4C). However, a parallel evaluation of selected gene expression in HT29 WT cells highlighted a key difference: the response to irradiation corresponded to PARP13 status in cells, but not to cell environment conditions. In contrast, irradiated DLD1 PARP13(−/−) cells exhibited a different expression pattern compared to HT29 cells (Additional Fig. 10A).
Furthermore, pathway analysis revealed a significant number of genes enriched in immune response pathways in a microenvironment dependent manner (Fig. 4B). Previously we have identified immune response as the most significantly altered in HT29 WT cells after multifractionated irradiation [22], therefore we next investigated a possible impact of PARP13 in regulating immune response genes. We compared the gene list from HT29 PARP13(−/−) microarray analysis with previous dataset obtained from HT29 wt cells cultured in 3D. Hierarchical heat map cluster analysis (Fig. 4D) revealed a striking difference in the immune related gene expression response to irradiation between HT29 wt and PARP13(−/−) cells. Wild type cells exhibited a strong activation of the selected genes whereas PARP13 KO cells displayed a diminished response with minimal expression changes in both 2D and 3D. These findings suggested that PARP13 may play a critical role in activating immune response upon ionizing radiation treatment under lr-ECM 3D conditions. To confirm the differential response of immune related genes in HT29 PARP13(−/−) cells upon irradiation (Fig. 4E), we next performed RT-qPCR analysis of selected IL29, OAS1, OAS2, IFITM1, IFIT1 genes. Notably, OAS1, OAS2, IFITM1 and IFIT1 did not respond to irradiation in PARP13(−/−) cells grown in either 2D or 3D, validating the impact of PARP13 on selected gene expression in CRC cells. On the other hand, a parallel investigation in irradiated DLD1 PARP13(−/−) cells exposed a cell type specific response indicating only a differential pattern of OAS2, IFITM1 and IFIT1 in cells grown in monolayer (Additional Fig. 10B).
Taken together, the microarray data revealed that PARP13 deficiency in HT29 cells significantly disrupts the activation of immune response signaling pathways upon multifractionated ionizing irradiation.
Chemoradiotherapy induces unconventional PARP expression in rectal cancer tissue
To evaluate the prognostic value of selected PARP gene expression levels in CRC, we collected paired tumor and adjacent normal tissue samples from rectal cancer patients both before and after their neoadjuvant chemo-radiotherapy (CRT) course (Fig. 5A). The expression levels of PARP9, PARP12, PARP13, and PARP14 were analyzed in the selected patient cohort using RT-qPCR (Fig. 5B, for fold change plots see Additional Fig. 11). Surprisingly, we found that the expression levels of all investigated PARPs were significantly increased in tumor tissue after CRT (p < 0.001 in all cases). To our delight, no statistically significant differences were observed in adjacent normal tissue in response to CRT, except the expression level of PARP12 was found to be slightly lower than before CRT (p < 0.05) signaling a significantly specific tumor cell PARP-related response to the therapy course.
The diagnostic sensitivity and specificity of selected PARPs expression levels before CRT were examined by ROC curve analysis (Additional Fig. 12A) demonstrating that PARP13 had the most significant prognostic accuracy (AUC = 0.901, p < 0.001). In addition, PARP13 expression was consistently higher in tumor than in adjacent normal tissues in TCGA colon and rectal adenocarcinoma datasets (Additional Fig. 13A). For the statistical analysis we stratified selected PARP expression levels (dCt) achieved before CRT and fold of expression after CRT (ddCt) into high and low based on the average value in each group. We then explored the distribution of low and high fold of expression after CRT of PARP9, PARP12, PARP13, and PARP14 in both tumor and adjacent normal tissue sample groups from rectal cancer patients. To achieve this, we applied Chi-square or Fisher’s exact tests to identify associations between changes in expression and demographic and clinicopathological characteristics (see Additional Tables 8 and 9). Although changes in PARP12 expression in tumor tissue were significantly associated with lymph node status after CRT, no other statistically significant PARP associations with patient variables were found.
Next, we assessed the prognostic value of PARP9, PARP12, PARP13 and PARP14 expression in rectal cancer tumor tissues using Kaplan-Meier analysis, calculating the time of overall survival (OS) and progression-free survival (PFS). Upon analyzing OS using PARPs expression levels before and after CRT (Additional Fig. 14), we found that prior to treatment only higher PARP9 expression levels are significantly linked to lower survival rates (p = 0.014). The analysis of OS in patients, stratified by changes in PARPs expression in tumor tissue induced by CRT (Fig. 5C), revealed a worse survival prognosis when there is a lower change in PARP9 and PARP14 expression (p = 0.025 in both cases). However, no statistically significant association was found between selected PARP expression and PFS rates in rectal cancer patients (Additional Fig. 12B). Additionally, analysis of TCGA data on association of PARP13 expression with overall and progression-free survival confirmed that PARP13 does not hold significant prognostic value for PARP13 in colorectal cancer (Additional Fig. 13B). Overall, these findings suggest that PARP9 and PARP14 expression profiles could be important prognostic factors for survival in rectal cancer patients.
Subsequently, we analyzed rectal cancer prognostic factors for OS using univariate and multivariate Cox regression analyses of tissue samples (Additional Table 10). The univariate Cox regression analysis (Additional Fig. 12C) identified primary tumor and lymph node status after treatment, along with Dworak tumor regression grade as prognostic factors for the OS. The final multivariate Cox regression analysis (Fig. 5D) uncovered that the change of PARP9 and PARP14 expression could be significant independent prognostic factors for overall survival (HR: 0.15, 95% CI: 0.02–0.88, p = 0.035 and HR: 0.07, 95% CI: 0.01–0.46, p = 0.005, respectively).
Given that multifractionated ionizing radiation induces the expression of immune response genes in vitro, we additionally explored whether the PARPs are co-expressed with selected OAS2, IFITM1, and IFIT1 genes in rectal cancer patients. The expression analysis (Additional Fig. 15A) showed significant downregulation of IFITM1 in tumor tissue, while IFIT1 was upregulated in both tumor and normal tissue after CRT. Additionally, we observed a lower expression level of OAS2 in normal tissue after CRT. Our co-expression analysis indicated a significant and strong positive correlation between the expression of all selected PARPs, OAS2 and IFIT1 in tumor tissue suggesting a coregulatory mechanism within the tumor microenvironment (Additional Fig. 16). In contrast, such PARP-immune gene co-expression was not evident in normal adjacent tissue samples displaying only a weak positive correlation between PARP14 and IFIT1 expression after CRT, suggesting a tumor tissue-specific association. While gene expression analysis revealed changes in tumor tissue, these did not translate into a significant diagnostic potential based on ROC analysis (Additional Fig. 15B). Finally, Kaplan-Meier and Cox regression analysis did not associate changes in the expression of these genes neither with overall nor progression-free survival of rectal cancer patients (Additional Fig. 17, Additional Table 11).
Taken together, this study demonstrates that CRT significantly increases the expression of PARP9, PARP12, PARP13 and PARP14, specifically in rectal tumor tissue. Notably, high pre-treatment PARP9 expression and a weaker post-treatment increase in PARP9 and PARP14 expression emerged as independent predictors of worse overall survival in rectal cancer patients. Although PARP13’s overall role in rectal cancer tissue was less evident, PARP13 was the most significant tumor tissue discriminator.
Discussion
Radiotherapy remains a cornerstone of cancer treatment, with advancements in delivery techniques and treatment significantly improving patient outcomes by minimizing toxicity to healthy tissues [31]. However, limitations persist due to intrinsic radiation resistance in some tumors, necessitating the development of more sophisticated strategies. Radiosensitizers hold promise in enhancing the efficacy of radiotherapy, and the DDR system presents a compelling target for biological therapy strategies [5]. ADP-ribosylation, a key protein modification in early DNA damage sensing, has been implicated in radiosensitization. Studies have shown that PARP inhibitors can increase tumor cell sensitivity to irradiation [10, 32, 33]. Building upon this established role of PARPs in DDR, we sought to broaden our understanding of understudied members of the PARP family, with the aim of identifying novel radiosensitizing agents for colorectal cancer treatment. We demonstrate a complex interplay between the expression of less studied PARPs, cellular microenvironment context and radiation response.
In contrast to monolayer, three-dimensional cell cultures more accurately recapitulate the in vivo tumor microenvironment, encompassing intercellular and extracellular interactions as well as gradients of nutrients and metabolites [34]. Moreover, the tumor microenvironment can have a profound effect on radiotherapy response as factors such as stromal cells interactions, ECM stiffness and mechanical forces might affect treatment efficacy [35]. We have previously shown that colorectal cancer cells grown in laminin-rich 3D culture exhibit clinically relevant gene expression profiles for the investigation of theranostic biomarkers in rectal cancer [23, 36]. Moreover, this experimental design incorporating clinically relevant radiation exposure could assist in discovering putative molecular pathways linked to radioresistance through the utilization of 3D cell culture models as well [22]. In the present study, we suggest that PARP family members exhibit diverse and context-dependent roles in the cellular response to irradiation. We observed a notable upregulation of PARP9, PARP12, PARP13 and PARP14 across three-dimensional cell culture models, including lr-ECM 3D and multicellular spheroids, following clinically relevant multifractionated irradiation. Intriguingly, the observed PARP expression pattern might be unique to cancer cells, as no corresponding expression changes were found in normal cells following irradiation suggesting cancer-specific therapeutic adjuvant intervention (Fig. 1). Together these findings indicate that the radiosensitizing potential of unconventional PARPs should be tested using 3D cell cultures, which may provide a physiologically relevant platform for identification of novel therapeutic strategies.
Previous studies have consistently demonstrated that cells grown in 3D cultures exhibit increased radioresistance compared to 2D cultures [37–39]. Therefore, given the advantages of 3D cell culture in mimicking the in vivo tumor microenvironment, the applications of such cell models could profoundly implicate the development of innovative and effective radiotherapy strategies [40]. Our present findings highlight the radiosensitizing potential of understudied PARPs, with their effect dependent on both cancer cell type and cellular microenvironment (Fig. 2). PARP KO in DLD1 cells resulted in an elevated radiosensitivity cultivating them only in monolayer, while HT29 PARP KO cells remained susceptible to IR in 3D models. PARP13 emerged as the most critical gene for HT29 cell survival following radiation exposure including 3D cell culture settings. We discovered that HT29 cells lacking PARP13 display increased sensitivity to both single and multi-fractionated radiation regimens, resulting in impaired survival and a loss of spheroid integrity. Previous findings indicated that PARP13 knockdown significantly decreased HeLa cell viability via destabilization of TRAILR4 transcript, thereby sensitizing cells to apoptosis [30, 41], suggesting that the sensitivity to irradiation could be intrinsically related to the dysregulation of cell survival genes due to PARP13 knockout. In line with these observations, we showed that PARP13 deficiency resulted in the upregulation of TRAIL and TRAIL-R2/DR5 encoding genes, TNFSF10 and TNFRSF10B, respectively, which could drive the intrinsic susceptibility of cancer cells to apoptosis [42]. Alternatively, radiosensitization could be acquired due to disrupted regulation of IR-induced DDR response genes in PARP13 KO cells. However, the publicly available data on how PARP13 could impact cell survival in response to radiation is absent.
PARP13 is an RNA-binding protein that is primarily involved in the activation of cell antiviral immune response [43]. During viral infection, PARP13 destabilizes target RNA by direct binding and the recruitment of exosome factors to initiate their decay [44]. In addition, the translation of specific targets could be also repressed by binding of PARP13 to the eIF4A translation-initiation factor [45]. Regarding these findings, we subsequently revealed that PARP13 KO in HT29 cells resulted in a strong deregulation of RNA levels of genes involved in viral immune response pathways (Fig. 3), demonstrating that PARP13 could regulate host mRNA stability in CRC cells. This observation is in agreement with a recent report indicating that PARP13 could keep the immune response primed for stress stimulus in cell basal state [43]. Moreover, our findings show that PARP13 might influence the regulation of the PARP family itself. Knocking out PARP13 led to overexpression of PARP9, PARP10, PARP12 and PARP14. Despite PARP13 being catalytically inactive, it could be MARylated by PARP14, indicating the interplay among PARP family proteins [46]. Meanwhile, PARP14 activity is directly regulated by PARP9/DT3XL, enabling hydrolytic activity of PARP9 and DT3XL interaction with PARP14 [47]. Therefore, our findings illustrate a significant interaction between the respective PARPs existing at RNA levels as well.
In cancer cells, irradiation can promote the formation of micronuclei, whose nuclear envelope is prone to rupture, releasing DNA into cytosol. The exposed cytosolic DNA subsequently triggers IFN signaling cascade leading to the induced transcription of interferon-stimulated genes (ISGs) [48, 49]. Previously, we have showed that fractionated irradiation induced a significant overexpression of immune response genes including ISGs [22]. To our surprise, a subsequent microarray analysis revealed that PARP13 deficiency in HT29 cells significantly attenuated the activation of immune response pathways (Fig. 4), pointing at PARP13 as a key regulator of IFN pathway response to IR. Our findings show that IFITM1 and IFIT1 are among strongly induced in HT29 wild type cells but not in PARP13 knockout cells. Notably, both IFITM1 and IFIT1 are part of the interferon-related DNA damage resistance signature (IRDS) genes, which has been linked to the development of resistance to radiation therapy, chemotherapy, and immune checkpoint blockades in various cancers, and promotion tumor growth, metastasis and immune evasion [50, 51]. IFIT1 is frequently upregulated in DNA damage-resistant cancer cell sublines, including those resistant to radiation or chemotherapy [52] and residual tumors following treatment, such as in estrogen receptor–negative breast cancer following chemotherapy [53]. Similarly, IFITM1 expression is regulated by interferon-alpha/NF-κB signaling, and its suppression has been shown to reduce tumor growth and invasion in triple-negative breast cancer [54]. Together, these findings suggest that both IFIT1 and IFITM1 contribute to therapy resistance and tumor progression, likely through modulation of interferon and growth factor signaling pathways and may represent potential targets for therapeutic intervention in cancers characterized by high PARP13 and ISG expression. Moreover, PARP13 levels are found to be elevated upon IFN stimulation both in vitro and in vivo [55, 56], demonstrating its reciprocal regulation. The activation of ISGs can either stimulate intrinsic cancer cell DDR response to IR via STAT1 pathway [52, 57] or foster tumor immune cell evasion [58, 59] resulting in immunosuppressive tumor microenvironment. As a result, the elevated levels of ISGs are frequently associated with radioresistance in cancer cells [58, 60], proposing that PARP13 could maintain cell survival by regulating RNA stability of ISGs. Interestingly, we also detected an upregulated expression of CD274, encoding PD-L1, in irradiated PARP13 KO cells, suggesting a potential immunotherapeutic window for tumors with lower PARP13 expression.
Finally, we demonstrated that chemoradiotherapy significantly upregulates the expression of PARP9, PARP12, PARP13 and PARP14 exclusively within rectal tumor tissues (Fig. 5). This finding, observed solely in cancerous tissues, aligns with our in vitro cell culture results, suggesting a strong clinical relevance. Given these observations, prospective of these genes as therapy targets or therapy response biomarkers could be promising direction for future studies as identification of patients who are more likely to benefit from treatment could lead to more personalized therapy approaches and improved outcomes. Regarding PARP13, it emerged as the most significant discriminator between tumor and healthy tissues demonstrating a specific PARP13 role in cancerous cells. However, our data suggest that PARP13 contributes to tumor response to radiotherapy through indirect mechanisms, such as the regulation of other PARPs and immune response pathways, rather than serving as a standalone clinical biomarker. Surprisingly, our findings highlighted the potential clinical significance of PARP9 and PARP14 in rectal cancer. The high pre-treatment PARP9 levels and a blunted post-treatment increase in both PARP9 and PARP14 expression emerged as independent predictors of poorer overall survival in rectal cancer patients. The observed irradiation induced cross-regulation between PARP13 and PARP9 or PARP14 is particularly noteworthy, given that high pre-treatment PARP9 expression, and lower changes of both PARP9 and PARP14 expression post-CRT correlates with poorer survival outcomes. This indicates that PARP13 may indirectly modulate treatment response by regulating PARP9 and PARP14 related pathways, emphasizing the necessity for further investigation of PARPs in colorectal cancer. Our observations are supported by previous studies which demonstrated an elevated expression of PARP9 and PARP14 in many malignancies [61–66] whereas high levels of PARP14 are associated with poor prognosis in patients [67]. As a result, several PARP14 inhibitors are being recently utilized in preclinical development [68, 69] suggesting their potential adjuvant applicability in radiotherapy.
Study limitations
We recognize that the absence of direct protein-level validation in this study limits our ability to precisely map the downstream pathways and their contributions to irradiation response in colorectal cancer cells. Future studies should combine proteomic analyses with detailed functional assays in both wild-type and PARP13 KO HT29 to fully elucidate the role of PARP13 in modulating gene regulation, cell survival and therapy resistance in colorectal cancer models.
Conclusion
To summarize, we demonstrate that interferon-associated PARPs are significantly involved in CRC response to radiotherapy in preclinical and clinical settings. PARP13 emerged as the most crucial for CRC sensitivity to IR treatment in vitro, serving as a keystone regulating ISG immune response in both nonirradiated or IR treated cells, and is the most significant discriminator between tumor and healthy tissues. Simultaneously, albeit less evident in cell culture, elevated coexpression levels of PARP9 and PARP14 were associated with poor patient outcomes after chemoradiotherapy. Taken together, our results propose that a collective focus on the relationship between PARP9/13/14 axis proteins may promote synergistic therapeutic and theranostic outcomes for radiotherapy development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all current and former members of the Molecular Oncology laboratory for their support in this project. We thank Dr. Renatas Krasauskas for insightful advice on molecular cloning. Lastly, we thank all the patients for their participation in the study.
Abbreviations
- CRC
Colorectal cancer
- CRT
Chemoradiotherapy
- IR
Ionizing radiation
- DDR
DNA damage response
- SSB
Single stranded break
- DSB
Double stranded break
- ECM
Extracellular matrix
- lr-ECM 3D
Laminin rich extracellular matrix 3D cell culture
- MCS
Multicellular spheroid cell culture
- DEG
Differentially expressed gene
- KO
Knockout
- PARP
Poly(ADP-ribose) polymerase
- ISG
Interferon stimulated genes
- OS
Overall survival
- PFS
Progression-free survival
Author contributions
RP, KK, GG, EB, DN, VT, KV and DN carried out the experiments and managed all parts of the study. ES, AD, JV conducted sample collection. RP, VS, ES, AP and KS contributed to experimental design. RP and VS conducted data analyses and wrote the manuscript. KS, VS made critical revisions to the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the project “Programming cells and management of tumor microenvironment for personal therapy in oncology– LASTER” (No. VP1-3.1-SMM-10-V-02-027), grant No. S-LB-19-7 from the Research Council of Lithuania, grant No. MTEP-2 from the National Cancer Institute (Lithuania).
Data availability
The microarray datasets supporting the conclusions of this article are available in the GEO repository, GSE280331 and GSE280332.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Vilnius Region Biomedical Research (2017-07-04; No. of permission 158200-17-930-433) and informed consent was obtained from all participants. All clinical procedures were conducted at the National Cancer Institute in Lithuania between 2017 and 2022 according to Helsinki regulation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vaidotas Stankevicius, Email: vaidotas.stankevicius@gmc.vu.lt.
Kestutis Suziedelis, Email: kestutis.suziedelis@nvi.lt.
References
- 1.Ferlay J, Ervik M, Lam F, Laversanne M, Colombet M, Mery L et al. Global Cancer Observatory: Cancer Today. 2024. Cancer today. Available from: https://gco.iarc.who.int/today/
- 2.Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Translational Oncol. 2021;14(10):101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lievens Y, Borras JM, Grau C. Provision and use of radiotherapy in Europe. Mol Oncol. 2020;14(7):1461–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332(2):237–48. [DOI] [PubMed] [Google Scholar]
- 5.Huang RX, Zhou PK. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Sig Transduct Target Ther. 2020;5(1):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai P. Biology of Poly(ADP-Ribose) polymerases: the factotums of cell maintenance. Mol Cell. 2015;58(6):947–58. [DOI] [PubMed] [Google Scholar]
- 7.Langelier MF, Riccio AA, Pascal JM. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014;42(12):7762–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant HE, Schultz N, Thomas H, Parker K. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(6860):913–7. [DOI] [PubMed] [Google Scholar]
- 10.Kong Y, Xu C, Sun X, Sun H, Zhao X, He N, et al. BLM helicase Inhibition synergizes with PARP Inhibition to improve the radiosensitivity of Olaparib resistant non-small cell lung cancer cells by inhibiting homologous recombination repair. Cancer Biology Med. 2022;19(8):1150–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheikh H, Ryder D, Bateman A, Chalmers A, Jackson A. Radiotherapy and olaparib in combination for carcinoma of the oesophagus: A phase I study. ctRO. 2023;40. Available from: https://www.ctro.science/article/S2405-6308(23)00039-3/fulltext [DOI] [PMC free article] [PubMed]
- 12.Tuli R, Surmak AJ, Reyes J, Armour M, Hacker-Prietz A, Wong J, et al. Radiosensitization of pancreatic Cancer cells in vitro and in vivo through Poly (ADP-ribose) polymerase inhibition with ABT-888. Translational Oncol. 2014;7(3):439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto M, Takii R, Takaki E, Katiyar A, Nakato R, Shirahige K, et al. The HSF1–PARP13–PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis. Nat Commun. 2017;8(1):1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhoonmoon A, Nicolae CM, Moldovan GL. The KU-PARP14 axis differentially regulates DNA resection at stalled replication forks by MRE11 and EXO1. Nat Commun. 2022;13(1):5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhoonmoon A, Nicolae CM. Mono-ADP-ribosylation by PARP10 and PARP14 in genome stability. NAR Cancer. 2023;5(1):zcad009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicolae CM, Aho ER, Choe KN, Constantin D, Hu HJ, Lee D, et al. A novel role for the mono-ADP-ribosyltransferase PARP14/ARTD8 in promoting homologous recombination and protecting against replication stress. Nucleic Acids Res. 2015;43(6):3143–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan Q, Ding J, Khan SJ, Lawton LN, Shipp MA. DTX3L E3 ligase targets p53 for degradation at Poly ADP-ribose Polymerase-associated DNA damage sites. iScience. 2023;26(4):106444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catara G, Grimaldi G, Schembri L, Spano D, Turacchio G, Lo Monte M, et al. PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of golgi complex functions. Sci Rep. 2017;7:14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard IA, Burgess JT, O’Byrne KJ, Bolderson E. Beyond PARP1: the potential of other members of the Poly (ADP-Ribose) polymerase family in DNA repair and Cancer therapeutics. Front Cell Dev Biol. 2022;9:801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abuwatfa WH, Pitt WG, Husseini GA. Scaffold-based 3D cell culture models in cancer research. J Biomed Sci. 2024;31(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 2023;41(3):374–403. [DOI] [PubMed] [Google Scholar]
- 22.Stankevicius V, Vasauskas G, Rynkeviciene R, Venius J, Pasukoniene V, Aleknavicius E, et al. Microenvironment and Dose-Delivery-Dependent response after exposure to ionizing radiation in human colorectal Cancer cell lines. Radiat Res. 2017;188(3):291. [DOI] [PubMed] [Google Scholar]
- 23.Stankevicius V, Vasauskas G, Noreikiene R, Kuodyte K, Valius M, Suziedelis K. Extracellular Matrix-dependent pathways in colorectal Cancer cell lines reveal potential targets for anticancer therapies. AR. 2016;36(9):4559–68. [DOI] [PubMed] [Google Scholar]
- 24.Stankevicius V, Kunigenas L, Stankunas E, Kuodyte K, Strainiene E, Cicenas J, et al. The expression of cancer stem cell markers in human colorectal carcinoma cells in a microenvironment dependent manner. Biochem Biophys Res Commun. 2017;484(4):726–33. [DOI] [PubMed] [Google Scholar]
- 25.Benchling [Biology Software] [Internet]. 2023. Available from: https://benchling.com.
- 26.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elizarraras JM, Liao Y, Shi Z, Zhu Q, Pico AR, Zhang B. WebGestalt 2024: faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024;52(W1):W415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 29.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todorova T, Bock FJ, Chang P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun. 2014;5(1):5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra RA, Keane FK, Voncken FEM, Thomas CR. Contemporary radiotherapy: present and future. Lancet. 2021;398(10295):171–84. [DOI] [PubMed] [Google Scholar]
- 32.Sheikh H, Ryder D, Bateman A, Chalmers A, Jackson A. Radiotherapy and Olaparib in combination for carcinoma of the oesophagus: A phase I study. Clin Translational Radiation Oncol. 2023;40:100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuli R, Shiao SL, Nissen N, Tighiouart M, Kim S, Osipov A, et al. A phase 1 study of veliparib, a PARP-1/2 inhibitor, with gemcitabine and radiotherapy in locally advanced pancreatic cancer. eBioMedicine. 2019;40:375–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodrigues DB, Reis RL, Pirraco RP. Modelling the complex nature of the tumor microenvironment: 3D tumor spheroids as an evolving tool. J Biomed Sci. 2024;31(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antonelli F. 3D cell models in radiobiology: improving the predictive value of in vitro research. Int J Mol Sci. 2023;24(13):10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunigenas L, Stankevicius V, Dulskas A, Budginaite E, Alzbutas G, Stratilatovas E, et al. 3D cell Culture-Based global MiRNA expression analysis reveals miR-142-5p as a theranostic biomarker of rectal Cancer following neoadjuvant Long-Course treatment. Biomolecules. 2020;10(4):613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akolawala Q, Rovituso M, Versteeg HH, Rondon AMR, Accardo A. Evaluation of Proton-Induced DNA damage in 3D-Engineered glioblastoma microenvironments. ACS Appl Mater Interfaces. 2022;14(18):20778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Babel L, Grunewald M, Lehn R, Langhans M, Meckel T. Direct evidence for cell adhesion-mediated radioresistance (CAM-RR) on the level of individual integrin β1 clusters. Sci Rep. 2017;7(1):3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang Y, Liang S, Gao J, Wang Z, Li C, Wang R, et al. Extracellular matrix stiffness mediates radiosensitivity in a 3D nasopharyngeal carcinoma model. Cancer Cell Int. 2022;22(1):364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vehlow A, Deville SS, Cordes N. 3D Radiation Biology for Identifying Radiosensitizers. In: Willers H, Eke I, editors. Molecular Targeted Radiosensitizers: Opportunities and Challenges. Cham: Springer International Publishing; 2020. pp. 115–35. Available from: 10.1007/978-3-030-49701-9_6
- 41.Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun. 2013;4(1):2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shetty S, Gladden JB, Henson ES, Hu X, Villanueva J, Haney N, et al. Tumor necrosis factor-related apoptosis inducing ligand (TRAIL) up-regulates death receptor 5 (DR5) mediated by NFκB activation in epithelial derived cell lines. Apoptosis. 2002;7(5):413–20. [DOI] [PubMed] [Google Scholar]
- 43.Busa VF, Ando Y, Aigner S, Yee BA, Yeo GW, Leung AKL. Transcriptome regulation by PARP13 in basal and antiviral States in human cells. iScience. 2024;27(4):109251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwerk J, Soveg FW, Ryan AP, Thomas KR, Hatfield LD, Ozarkar S, et al. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat Immunol. 2019;20(12):1610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y, Wang X, Goff SP, Gao G. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J. 2012;31(21):4236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carter-O’Connell I, Vermehren-Schmaedick A, Jin H, Morgan RK, David LL, Cohen MS. Combining chemical genetics with Proximity-Dependent labeling reveals cellular targets of Poly(ADP-ribose) polymerase 14 (PARP14). ACS Chem Biol. 2018;13(10):2841–8. [DOI] [PubMed] [Google Scholar]
- 47.Kar P, Chatrin C, Đukić N, Suyari O, Schuller M, Zhu K et al. PARP14 and PARP9/DTX3L regulate interferon-induced ADP-ribosylation. EMBO J. 2024;1–25. [DOI] [PMC free article] [PubMed]
- 48.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15(7):405–14. [DOI] [PubMed] [Google Scholar]
- 49.Schneider WM, Chevillotte MD, Rice CM. Interferon-Stimulated genes: A complex web of host defenses. Annu Rev Immunol. 2014;32(1):513–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace TA, Martin DN, Ambs S. Interactions among genes, tumor biology and the environment in cancer health disparities: examining the evidence on a National and global scale. Carcinogenesis. 2011;32(8):1107–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, et al. IFNβ‐dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32(20):2751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DSA, Baker SW, Khodarev N, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci. 2008;105(47):18490–5. [DOI] [PMC free article] [PubMed]
- 53.Legrier ME, Bièche I, Gaston J, Beurdeley A, Yvonnet V, Déas O, et al. Activation of IFN/STAT1 signalling predicts response to chemotherapy in oestrogen receptor-negative breast cancer. Br J Cancer. 2016;114(2):177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Provance OK, Geanes ES, Lui AJ, Roy A, Holloran SM, Gunewardena S, et al. Disrupting interferon-alpha and NF-kappaB crosstalk suppresses IFITM1 expression attenuating triple-negative breast cancer progression. Cancer Lett. 2021;514:12–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin YT, Chiweshe S, McCormick D, Raper A, Wickenhagen A, DeFillipis V, et al. Human cytomegalovirus evades ZAP detection by suppressing CpG dinucleotides in the major immediate early 1 gene. PLoS Pathog. 2020;16(9):e1008844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li MMH, Aguilar EG, Michailidis E, Pabon J, Park P, Wu X, et al. Characterization of novel splice variants of zinc finger antiviral protein (ZAP). J Virol. 2019;93(18):e00715–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in transduced sensitive cells. Proceedings of the National Academy of Sciences. 2004;101(6):1714–9. [DOI] [PMC free article] [PubMed]
- 58.Post AEM, Smid M, Nagelkerke A, Martens JWM, Bussink J, Sweep FCGJ, et al. Interferon-Stimulated genes are involved in Cross-resistance to radiotherapy in Tamoxifen-Resistant breast Cancer. Clin Cancer Res. 2018;24(14):3397–408. [DOI] [PubMed] [Google Scholar]
- 59.Chen RH, Xiao ZW, Yan XQ, Han P, Liang FY, Wang JY, et al. Tumor cell-Secreted ISG15 promotes tumor cell migration and immune suppression by inducing the macrophage M2-Like phenotype. Front Immunol. 2020;11:594775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goedegebuure RSA, Vonk C, Kooij LP, Derks S, Thijssen VLJL. Combining radiation therapy with interferons: back to the future. Int J Radiation Oncology*Biology*Physics. 2020;108(1):56–69. [DOI] [PubMed] [Google Scholar]
- 61.Kamata T, Paschal B, Kamata T, Paschal B. ADP-Ribosylation of the Ubiquitin C-Terminus by Dtx3L/Parp9. In: Ubiquitin Proteasome System - Current Insights into Mechanism Cellular Regulation and Disease. IntechOpen; 2018. Available from: https://www.intechopen.com/chapters/64167
- 62.Tang X, Zhang H, Long Y, Hua H, Jiang Y, Jing J. PARP9 is overexpressed in human breast cancer and promotes cancer cell migration. Oncol Lett. 2018;16(3):4073–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang CS, Jividen K, Spencer A, Dworak N, Ni L, Oostdyk LT, et al. Ubiquitin modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol Cell. 2017;66(4):503–e5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbarulo A, Iansante V, Chaidos A, Naresh K, Rahemtulla A, Franzoso G, et al. Poly(ADP-ribose) polymerase family member 14 (PARP14) is a novel effector of the JNK2-dependent pro-survival signal in multiple myeloma. Oncogene. 2013;32(36):4231–42. [DOI] [PubMed] [Google Scholar]
- 65.Iansante V, Choy PM, Fung SW, Liu Y, Chai JG, Dyson J, et al. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat Commun. 2015;6(1):7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao N, Chen Q, Shi W, Tang L, Fu Y. PARP14 promotes the proliferation and gemcitabine chemoresistance of pancreatic cancer cells through activation of NF-κB pathway. Mol Carcinog. 2019;58(7):1291–302. [DOI] [PubMed] [Google Scholar]
- 67.Zhang L, Chen W, Shi Z, Shang Z. PARP14 correlates with GBM proliferation and poor prognosis by elevating expression of SAMD/SAMD9L. Ir J Med Sci. 2024;193(2):585–93. [DOI] [PubMed] [Google Scholar]
- 68.Schenkel LB, Molina JR, Swinger KK, Abo R, Blackwell DJ, Lu AZ, et al. A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants. Cell Chem Biol. 2021;28(8):1158–e116813. [DOI] [PubMed] [Google Scholar]
- 69.Wigle TJ, Ren Y, Molina JR, Blackwell DJ, Schenkel LB, Swinger KK, et al. Targeted degradation of PARP14 using a heterobifunctional small molecule. ChemBioChem. 2021;22(12):2107–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray datasets supporting the conclusions of this article are available in the GEO repository, GSE280331 and GSE280332.