Abstract
Although a small fraction of human cancers have increased rates of somatic mutation because of known deficiencies in DNA repair, little is known about the prevalence of somatic alterations in the vast majority of human cancers. To systematically assess nonsynonymous somatic alterations in colorectal neoplasia, we used DNA sequencing to analyze ≈3.2 Mb of coding tumor DNA comprising 1,811 exons from 470 genes. In total, we identified only three distinct somatic mutations, comprising two missense changes and one 14-bp deletion, each in a different gene. The accumulation of approximately one nonsynonymous somatic change per Mb of tumor DNA is consistent with a rate of mutation in tumor cells that is similar to that of normal cells. These data suggest that most sporadic colorectal cancers do not display a mutator phenotype at the nucleotide level. They also have significant implications for the interpretation of somatic mutations in candidate tumor-suppressor genes.
Cancer results from the sequential accumulation of mutations in specific genes. As new mutations provide a growth advantage to a cell within the tumor, the selected clone outgrows the precursor lesion, and successive waves of mutation, clonal expansion, and growth lead to cancer progression. Throughout this process of mutation and selection, the acquisition of new mutations is thought to be the rate-limiting step, setting the timeline of tumor development. Although an increase in the rate of mutation should accelerate this process, there has been debate as to whether genetic instability or a “mutator phenotype” (1) in addition to selection is actually required for neoplasia. Some have argued that an increased mutation rate at the nucleotide level is necessary to account for all of the mutations present in cancers (1), while others have suggested that the normal somatic mutation rate, coupled with selection, is sufficient to explain tumor formation in most cases (2). Also, it has been suggested that a different form of genetic instability, involving gross changes in chromosomes rather than subtle changes in nucleotide sequence, drives most human cancers (1, 3, 4).
Recent work has demonstrated that a small fraction of cancers clearly display genetic instability at the nucleotide level because of defects in DNA repair processes. The first demonstration of this principle was made through the discovery that patients with xeroderma pigmentosum (XP) and related disorders have hereditary defects in genes encoding nucleotide excision repair enzymes (5). These patients develop skin tumors as a result of their inability to repair mutations caused by UV irradiation. XP and related disorders are inherited in an autosomal recessive fashion and are accordingly rare. Another form of repair defect, involving mismatch repair, has been shown to be implicated in the tumors of all patients with hereditary nonpolyposis colon cancer (HNPCC) syndrome (6), as well as ≈12% of patients with nonfamilial forms of colorectal cancer (7). HNPCC is inherited in an autosomal dominant fashion and accounts for 3–5% of the total colorectal cancer cases in the Western world (6, 8). The mismatch repair defects are predominantly due to inactivation of the hMSH2 or hMLH1 genes, although defects in other mismatch repair genes play a role in a subset of HNPCC cases. Although mismatch repair defects have been shown to lead to a widespread nucleotide instability, microsatellite sequences are particularly susceptible to this form of repair defect (9, 10). Accordingly, microsatellite instability (MIN), defined by insertion and deletion errors in repetitive elements, has been shown to occur in all colorectal tumors harboring mismatch repair defects. MIN also has been found in a small fraction of other cancers, including those of the stomach, endometrium, and ovary.
Unlike the aforementioned examples, the vast majority of human cancers seem to possess intact mismatch repair and base excision repair capabilities, and it is unclear whether they possess a mutator phenotype at the nucleotide level. Some studies have suggested an increase in the number of point mutations in tumors, for example, by analysis of published data on multiple mutations in the p53 gene (11), but such measurements have been limited to individual genes and may be compromised by a number of experimental variables. Other studies have suggested an elevated rate of somatic mutation in colorectal tumors on the basis of the analysis of sequences adjacent to dinucleotide repeats (12). To obtain an unbiased measurement of the prevalence of nucleotide alterations, we surveyed a large number of coding regions in the genomes of colorectal cancers. Our results, described below, have significant implications for theories relating instability to naturally occurring human tumorigenesis and the interpretation of somatic mutations in specific genes in such cancers.
Material and Methods
Collection of Gene Sequences.
Gene sequences were obtained from the Celera Discovery System (http://cds.celera.com) and GenBank (http://www.ncbi.nlm.nih.gov) databases. Determination of coding sequence within each transcript was based on annotation present in the databases. In each case, we used computational approaches to confirm start and stop codons and ORFs.
Primer Design and Oligonucleotide Synthesis.
Exon sequences and adjacent intronic sequences for each gene were extracted from Celera Discovery System or from GenBank databases. Primers for PCR amplification and DNA sequencing were designed by using the PRIMER 3 program (13) (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Forward primers were localized to regions no closer than 100 bp from the exon boundaries; forward sequencing primers were designed so that they would not overlap forward PCR primers and to extend no closer than 50 bp to the 5′ exon boundary. Reverse amplification primers were localized to regions no closer than 10 bp to the 3′ exon boundary. Oligonucleotides for PCR amplification and sequencing were synthesized by Genelink, NY.
Isolation of Tumor DNA.
Because primary tumors always contain nonneoplastic cells, presumptive mutations can be masked or difficult to interpret. To circumvent this difficulty, we analyzed early passage cell lines passaged in vitro or in nude mice, in which sequence changes were unambiguous. It has been shown previously that the genetic alterations in such cell lines usually are indistinguishable from those found in the primary tumors (14–16). Surgically removed colorectal tumors were disaggregated and implanted into nude mice or into in vitro culture conditions, as described (14). DNA was purified by standard SDS-proteinase K digestion and phenol-chloroform extraction or by DNAEasy Tissue kit (Qiagen, Chatsworth, CA).
Determination of MIN Status in Tumor Samples.
A large number of dinucleotide or tetranucleotide microsatellite sequences (>20) and the BAT26 mononucleotide marker (17) were evaluated in each of the tumors used in this cohort. PCR products were separated and analyzed on either manual sequencing gels or by using an SCE-9610 96-well capillary electrophoresis system (SpectruMedix, State College, PA).
PCR Amplification.
Each PCR was performed in 10 μl containing 0.25 mM dNTPs, 1 μM forward and reverse primers, 6% DMSO, 1× PCR buffer (Invitrogen), 5 ng DNA, and 0.5 units platinum Taq (Invitrogen). PCR was carried out in 384-well format (Applied Biosystems) with a touchdown PCR program: 94°C (2 min); 3 cycles of 94°C (30 sec), 64°C (30 sec), 70°C (30 sec); 3 cycles of 94°C (30 sec), 61°C (30 sec), 70°C (30 sec); 3 cycles of 94°C (30 sec), 58°C (30 sec), 70°C (30 sec); 35 cycles of 94°C for (30 sec), 57°C (30 sec), 70°C (30 sec); 1 cycle of 70°C (5 min). After PCR, samples were purified by isopropanol precipitation and resuspended in TE (10 mM Tris, pH 7.4/1 mM EDTA).
DNA Sequencing.
Purified PCR products were sequenced by using Big Dye Terminator v.3.0 Kit (Applied Biosystems). Cycle sequencing reactions were carried out in 384-well format at 35 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Sequencing reactions were precipitated by mixing with addition of a 20-μl mixture containing 87.5% ethanol and 0.112 M sodium acetate. The precipitated products were resuspended in sample loading buffer (Hi-Di Formamide, Applied Biosystems) and were analyzed with a SCE-9610 96-well capillary electrophoresis system (SpectruMedix).
Sequencing Data Analysis.
Sequencing data were analyzed by using BASECALL v.3.39 (SpectruMedix). SEQMAN (DNAstar, Madison, WI) and SEQUENCHER (Gene Codes, Ann Arbor, MI) were used to assemble sequence chromatograms and consensus reference sequences. Assemblies were visually inspected to identify potential alterations. Coding sequences containing nonsynonymous alterations were searched against Celera Discovery System RefSNP database to determine if the changes corresponded to previously identified polymorphisms.
Statistical Analysis.
The interval estimate for somatic mutations per Mb tumor DNA represents a “highest posterior density region” based on an exact a posteriori analysis, assuming a Poisson model for the distribution of mutations on the genome, and a uniform a priori distribution on the unknown mutation rate (18). It is preferable to standard asymptotic confidence intervals because there are few mutations and the likelihood function is skewed. Additional details on these analyses and statistical tools for evaluating whether observed mutation frequencies are above the background mutation frequency are available at http://astor.som.jhmi.edu/∼gp/trab/.
Results and Discussion
To explore the frequency of somatic alterations in coding regions of tumor DNA, we selected a total of 504 genes for analysis. By using a combination of the public (19) and private (Celera; ref. 20) genome databases, we extracted available genomic data for a total of 2,206 exons contained in these genes and designed primers for PCR amplification and sequencing. The sequences of these genes were determined in a panel of 12 colorectal cancer cell lines. Cell lines rather than primary tumors were used to ensure that nonneoplastic cells within the tumors did not complicate the sequence analysis. These cell lines were generally of early passage, and each was extensively examined with several different kinds of microsatellites to ensure that no MIN was present, thus excluding mismatch repair deficiency as a source of potential somatic alterations in these cases. The cell lines also were chosen to contain regions of loss of heterozygosity (LOH) in areas containing genes of interest to increase the sensitivity of detecting somatic mutations by DNA sequencing, as any alteration observed would appear as a homozygous change. The exons described above were amplified from tumor DNA samples using PCR and were subjected to dideoxy sequencing. A total of 1,811 exons were successfully amplified from 470 genes and sequenced from an average of 11 different tumor DNA samples (Table 1). The average size of examined exons was 150 bp and ranged up to 699 bp in length. Both intronic splice acceptor and donor regions were analyzed, with the exception of cases where the exon size was greater than the length of readable sequencing product. A total of ≈6.5 Mb of sequence data was obtained in this way (Table 1).
Table 1.
Summary of sequence analysis and observed alterations
| Selected genes | 504 |
| Exons extracted | 2,206 |
| Genes analyzed | 470 |
| Exons analyzed | 1,811 |
| Average exon size | 152 bp |
| Average number of tumors analyzed per exon | 11.4 |
| Total sequence data obtained | 6.5 Mb |
| Total coding DNA analyzed | 3.2 Mb |
| Nonsynonymous changes observed | 320 |
| Previously identified SNPs | 90 |
| Novel coding SNPs | 227 |
| Somatic mutations | 3 |
Sequencing results were analyzed by assembling data of related exons from different tumors and comparing them in silico to reference sequences (Fig. 1). We confined our analysis to 3.2 Mb that encoded exons and focused on protein altering (nonsynonymous) changes, as these provided the best indicators of alterations most likely to have an effect on tumorigenesis. A total of 320 alterations resulting in nonsynonymous changes were identified in the 3.2-Mb assembly. Observed changes first were compared with polymorphism databases to determine whether they corresponded to previously recognized single nucleotide polymorphisms (SNPs). A total of 90 SNPs were identified and excluded in this manner. The remaining changes, representing potential somatic alterations, then were analyzed by sequencing of DNA from normal tissues of the corresponding patients. Two hundred and twenty seven of these changes were present in the corresponding patient's normal tissues. These changes thus represented previously unidentified SNPs within exons.
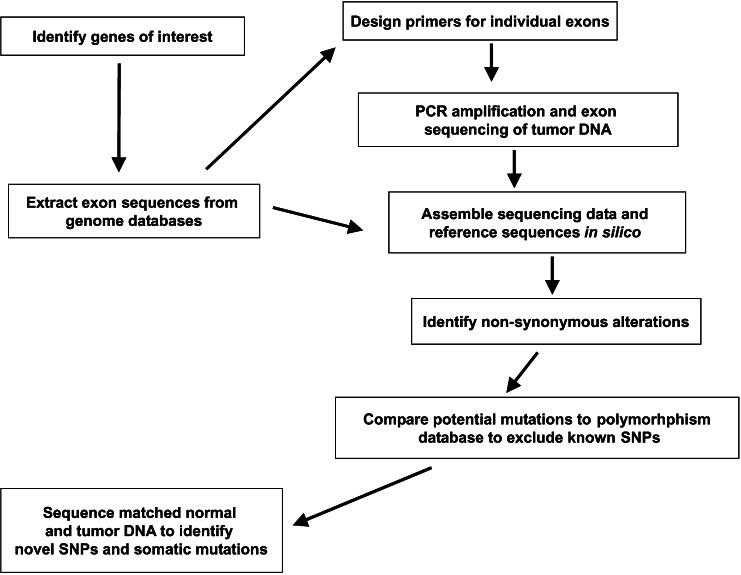
Figure 1.
Approach to somatic mutation discovery. After identifying genes of interest, the Celera and public genome databases were used to extract exon sequences and intronic regions to design primers for PCR amplification and sequencing. Successfully amplified and sequenced exons were assembled and compared with exon reference sequences to identify nonsynonymous alterations. Any potential alterations were then compared with polymorphism databases to exclude known SNPs. For remaining alterations, PCR products from matched normal and tumor DNA were sequenced to identify novel SNPs and somatic mutations.
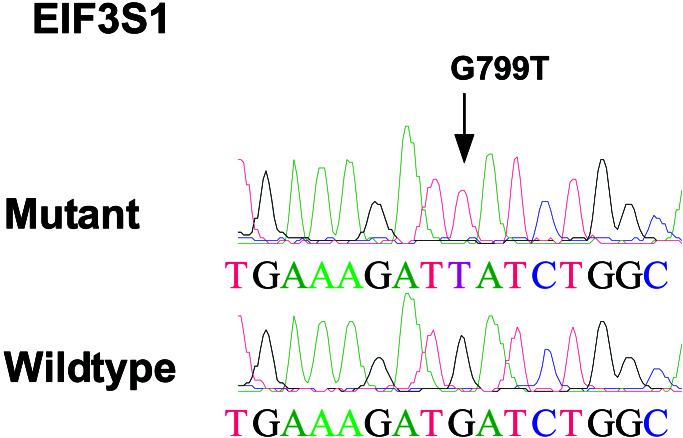
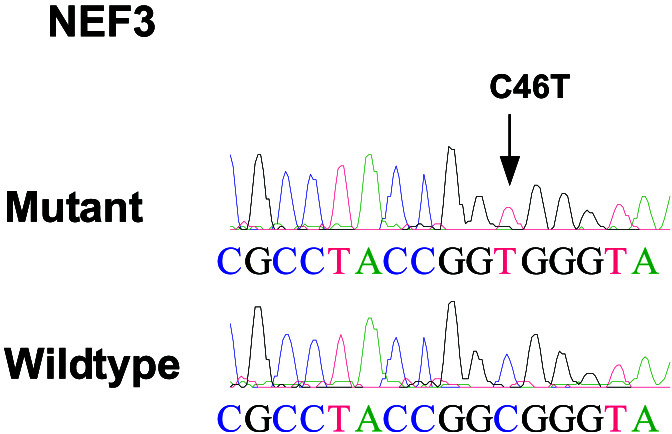
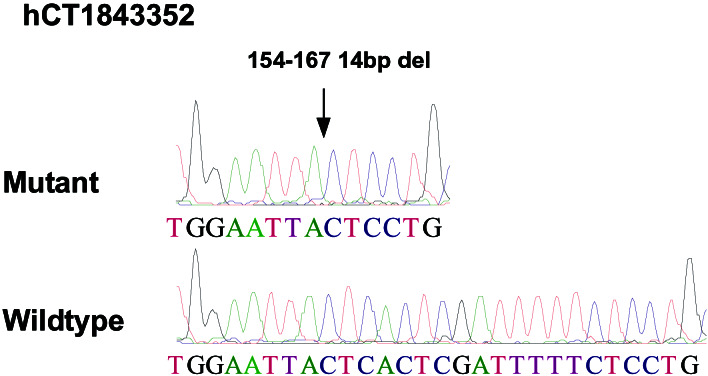
After analysis of all potential changes by sequencing of both tumor and matched normal DNA samples, only three somatic alterations were identified, each in a different gene (Table 2). Two of these corresponded to missense changes in well characterized genes: a G to T transversion at nucleotide position 799 of the eukaryotic translation initiation factor 3 subunit 1 gene, resulting in an aspartate to tyrosine change at amino acid position 267 (Fig. 2), and a C to T transition at nucleotide position 46 of the neurofilament 3 gene, resulting in an arginine to tryptophan change at amino acid position 16 (Fig. 3). The third alteration corresponded to a 14-bp deletion in a gene of unknown function (Celera hCT1843352) at nucleotide position 154, resulting in a frameshift and introduction of premature translational termination codon at amino acid position 54 (Fig. 4). All three changes occurred in different tumor samples, and no mutations in the remaining exons of these genes were detected in the ≈10 additional tumors examined.
Table 2.
Genes identified with somatic alterations
| Gene name | Gene symbol | Celera transcript ID | GenBank accession | Nucleotide change* | Amino acid change |
|---|---|---|---|---|---|
| Eukaryotic translation initiation factor 3, subunit 1 | EIF3S1 | hCT30575 | NM_003758 | 799 G > T | 267 D > Y |
| Neurofilament 3 (150 kD medium) | NEF3 | hCT7649 | NM_005382 | 46 C > T | 16 R > W |
| N/A | N/A | hCT1843352 | AA121909† | 154 14-bp deletion | Frameshift and stop at aa 59 |
Nucleotide positions are relative to the transcript sequences present in the Celera Discovery System.
Represents an unspliced expressed sequence tag that is homologous to one exon of the Celera predicted transcript.
Figure 2.
Sequence chromatograms showing somatic alteration in eukaryotic translation initiation factor 3, subunit 1. Arrow indicates the location of nucleotide change in the tumor DNA sequence.
Figure 3.
Sequence chromatograms showing somatic alteration in neurofilament 3 (150 kD medium). Arrow indicates the location of nucleotide change in the tumor DNA sequence.
Figure 4.
Sequence chromatograms showing somatic alteration in Celera predicted transcript hCT1843352. Arrow indicates the region of the 14-bp deletion in the tumor DNA sequence.
The identification of three somatic alterations from 3.2 Mb of analyzed DNA corresponds to a somatic mutation frequency of approximately one alteration (0.22–2.5 alterations, 95% probability interval) per Mb of tumor DNA, or approximately 3,000 alterations (668–7450 alterations, 95% probability interval) per haploid genome. However, this represents an underestimate of the number of somatic changes in a tumor genome, as we examined only nonsynonymous changes. On the basis of genome-wide polymorphism studies (21, 22), it has been shown that nonsynonymous and synonymous changes have similar frequencies in coding DNA, and that the frequency of coding and noncoding changes is approximately the same. Therefore, one would expect that our somatic mutation frequency would be increased approximately 2-fold. This probably represents the higher end of such an estimate, because approximately two-thirds of random coding mutations alter an amino acid, and the selection pressure on these nonsynonymous changes is likely to be substantially less at the single-cell level than at the organismal level. Accordingly, the majority of cancers would be expected to carry less than 6,000 alterations per genome.
The total somatic alterations in a cancer represent those that accumulated during normal somatic cell division as well as in the tumor during successive waves of clonal expansion and growth. Each of the bottlenecks that the tumor cell passes through (including initiation plus each wave of clonal expansion) leads to fixation of all mutations that have previously occurred in the tumor cell's progenitors. Colorectal cancers are thought to develop from epithelial stem cells residing in colonic crypts (23–25). As the entire intestinal epithelium replaces itself every few days, a typical colon epithelial stem cell is thought to undergo >100 cell divisions per year and >4,000 divisions before the onset of most adenomas at age 40. Additionally, as cell turnover far exceeds net tumor growth (2, 26, 27), during the 20-yr progression of a colorectal carcinoma, at least another 2,000 cell divisions would occur. By using the estimated mutation rate in normal somatic human cells of 1 × 10−10–1 × 10−9 nucleotides per cell per division (1, 28, 29), one would expect that a 20-year old tumor removed from a 60-year old individual would have accumulated 1,800–18,000 alterations per haploid genome. This number of mutations is similar to the prevalence of alterations we observed in sporadic cancers and suggests that normal mutation rates are sufficient to explain tumor progression in most cancers.
Like all large-scale studies, our approach has several limitations. First, the majority of genes analyzed, including the three that contained somatic alterations, were present in regions that had undergone LOH in the tumor DNA. Although there is no current evidence to suggest a different rate of point mutations in regions of LOH, it is possible that our conclusions would be altered by analyses of additional genes from heterozygous regions. Second, our analysis was performed on cell lines rather than primary tumors, which may allow for accumulation of mutations during in vitro passaging. However, all of the mutations observed were clonal in the tumors analyzed, making it unlikely that they developed during the few in vitro passages of the cell lines. This possibility, even if true, would only decrease the actual number of mutations calculated to be present in the tumors and, therefore, would not impact on the major conclusions of this study. Third, if we assume a hierarchical model of epithelial cell generation in colonic crypts rather than the conventional single stem cell model (30), only ≈50 instead of 4,000 cell divisions would occur in an average tumor progenitor cell, and the number of expected mutations per tumor genome would be reduced from a range of 1,800–18,000 to 600–6,000. Although the current experimental evidence does not support such a model (25), the reduced number of mutations remains within several-fold of our observed mutation rate within tumors and would suggest at best a weak increase in mutation rates in cancer cells.
Despite these potential limitations, it is clear that these results have several significant implications. One implication is that they do not disprove the argument that genetic instability is an inherent feature of tumorigenesis. Instead, they emphasize that, although a subtle instability at the nucleotide level does not play a role in most colorectal cancers, a different form of genetic instability generally occurs in these tumors. In fact, extensive studies of polymorphic markers in our panel of tumors showed that they each had numerous allelic losses, consistent with widespread chromosomal changes (31). Additionally, the results highlight the importance of specific mutations in the genes that drive neoplasia. Because the studies described above show that passenger mutations (i.e., random mutations, with no functional significance) are found so rarely in these tumors, great confidence can be placed in the significance of somatic mutations in genes like APC and p53, which occur in nearly all of the colorectal cancers represented in our panel. Lastly, the results are important for interpreting the significance of mutations in candidate tumor-suppressor genes. The search for other tumor-suppressor genes involved in common cancers continues to be active, and the best evidence to validate such candidates comes from mutational data (32). On the one hand, our data show that mutations in any given gene are likely to be uncommon unless selected for during tumorigenesis. On the other hand, even random genes like those studied here are occasionally mutated in these cancers as a result of normal mutational processes and bottlenecks during the neoplastic process. A prudent conclusion from these results, therefore, would be to invoke the requirement for a significantly higher mutation frequency—for example, at least 2 independent examples of functionally altering mutations from a panel of no more than 20 tumors—to implicate a gene as a candidate tumor suppressor. To help investigators rigorously determine whether the number of observed mutations in a gene of interest is significantly above the expected background mutation frequency, we have developed simple statistical tools, freely available at http://astor.som.jhmi.edu/∼gp/trab/.
Acknowledgments
We thank members of the Molecular Genetics Laboratory for helpful discussions. This work was supported by the Maryland Cigarette Restitution Fund, the Clayton Fund, and National Institutes of Health Grants CA62924, CA57345, and CA43460.
Abbreviations
- XP
xeroderma pigmentosum
- MIN
microsatellite instability
- SNP
single nucleotide polymorphism
References
- 1.Loeb L A. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]
- 2.Tomlinson I, Bodmer W. Nat Med. 1999;5:11–12. doi: 10.1038/4687. [DOI] [PubMed] [Google Scholar]
- 3.Boland C R, Ricciardiello L. Proc Natl Acad Sci USA. 1999;96:14675–14677. doi: 10.1073/pnas.96.26.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 5.Bootsma D, Kraemer K H, Cleaver J E, Hoeijmakers J H J. In: The Genetic Basis of Human Cancer, Vol. 1. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 245–274. [Google Scholar]
- 6.Lynch H T, de La Chapelle A. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 7.Perucho M. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 8.Boland R C. In: The Genetic Basis of Human Cancer, Vol. 1. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 333–346. [Google Scholar]
- 9.Sia E A, Jinks-Robertson S, Petes T D. Mutat Res. 1997;383:61–70. doi: 10.1016/s0921-8777(96)00046-8. [DOI] [PubMed] [Google Scholar]
- 10.Thomas D C, Umar A, Kunkel T A. Mutat Res. 1996;350:201–205. doi: 10.1016/0027-5107(95)00112-3. [DOI] [PubMed] [Google Scholar]
- 11.Strauss B S. Carcinogenesis. 1997;18:1445–1452. doi: 10.1093/carcin/18.8.1445. [DOI] [PubMed] [Google Scholar]
- 12.Stoler D L, Chen N, Basik M, Kahlenberg M S, Rodriguez-Bigas M A, Petrelli N J, Anderson G R. Proc Natl Acad Sci USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozen S, Skaletsky H. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 14.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Willson J K, Markowitz S, Hamilton S R, Kern S E, et al. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 15.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 16.Cahill D P, Lengauer C, Yu J, Riggins G J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 17.Parsons R, Myeroff L L, Liu B, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 18.Schervish M. Statistical Theory. New York: Springer; 1995. [Google Scholar]
- 19.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature (London) 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 21.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane C R, Lim E P, Kalyanaraman N, et al. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 22.Halushka M K, Fan J B, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- 23.Lipkin M, Bell B, Shelrock P. J Clin Invest. 1963;42:767. doi: 10.1172/JCI104769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shorter R G, Moertel C G, Titus J L. Am J Dig Dis. 1964;9:760. doi: 10.1007/BF02231983. [DOI] [PubMed] [Google Scholar]
- 25.Potten C S, Loeffler M. Development (Cambridge, UK) 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 26.Matsui T, Yao T, Iwashita A. World J Surg. 2000;24:1022–1028. doi: 10.1007/s002680010153. [DOI] [PubMed] [Google Scholar]
- 27.Shimomatsuya T, Tanigawa N, Muraoka R. Jpn J Cancer Res. 1991;82:357–362. doi: 10.1111/j.1349-7006.1991.tb01854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drake J W. Nature (London) 1969;221:1128–1132. [PubMed] [Google Scholar]
- 29.Elmore E, Kakunaga T, Barrett J C. Cancer Res. 1983;43:1650–1655. [PubMed] [Google Scholar]
- 30.Morris J A. J Theor Biol. 1999;199:87–95. doi: 10.1006/jtbi.1999.0943. [DOI] [PubMed] [Google Scholar]
- 31.Thiagalingam S, Laken S, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B, Lengauer C. Proc Natl Acad Sci USA. 2001;98:2698–2702. doi: 10.1073/pnas.051625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haber D, Harlow E. Nat Genet. 1997;16:320–322. doi: 10.1038/ng0897-320. [DOI] [PubMed] [Google Scholar]