Abstract
Despite recent treatment advances, the majority of patients with chronic hepatitis C fail to respond to antiviral therapy. Although the genetic basis for this resistance is unknown, accumulated evidence suggests that changes in the heterogeneous viral population (quasispecies) may be an important determinant of viral persistence and response to therapy. Sequences within hepatitis C virus (HCV) envelope 1 and envelope 2 genes, inclusive of the hypervariable region 1, were analyzed in parallel with the level of viral replication in serial serum samples obtained from 23 patients who exhibited different patterns of response to therapy and from untreated controls. Our study provides evidence that although the viral diversity before treatment does not predict the response to treatment, the early emergence and dominance of a single viral variant distinguishes patients who will have a sustained therapeutic response from those who subsequently will experience a breakthrough or relapse. A dramatic reduction in genetic diversity leading to an increasingly homogeneous viral population was a consistent feature associated with viral clearance in sustained responders and was independent of HCV genotype. The persistence of variants present before treatment in patients who fail to respond or who experience a breakthrough during therapy strongly suggests the preexistence of viral strains with inherent resistance to IFN. Thus, the study of the evolution of the HCV quasispecies provides prognostic information as early as the first 2 weeks after starting therapy and opens perspectives for elucidating the mechanisms of treatment failure in chronic hepatitis C.
Infection with hepatitis C virus (HCV) is a leading cause of chronic liver disease worldwide (1). Although a broadly protective hepatitis C vaccine would be highly desirable, its development has proven to be exceedingly difficult (2). Thus, effective treatment for this disease is especially important. However, despite recent successes after the introduction of combination therapy with IFN-α and ribavirin (3–5), about 60% of patients still fail to respond. Therefore, resistance to antiviral therapy remains a serious problem in the management of chronic hepatitis C. Patients who do not have a sustained response to IFN therapy constitute a heterogeneous group (6): some experience persistent viremia and alanine aminotransferase abnormalities (nonresponse) whereas others have an initial response followed by reactivation while on IFN therapy (breakthrough) or by relapse after its discontinuation (transient response). Several studies have pointed to the HCV genotype and the baseline level of viremia as the most important predictive factors of responsiveness to IFN therapy (7). However, diverse treatment responses in patients with the same genotype and similar levels of viremia suggest that other factors may be responsible for the effectiveness of therapy.
Although the biological conditions associated with treatment failure are not presently known, both host and viral factors likely are involved. Among the latter, viral resistance to IFN is considered to play a major role, although the molecular basis of such resistance has not been fully defined (8–10). A specific region of the nonstructural 5A gene (11) has been suggested as a possible determinant of IFN sensitivity (12, 13), but this hypothesis is still highly controversial (14–19). More recently, a second region of potential interest has been identified within the envelope 2 glycoprotein (E2) (20), but its role in determining IFN resistance also remains unclear (21–24). Regardless of the precise genetic determinant(s) of the resistance of HCV to IFN, evidence accumulated over the past few years suggests that the genetic variability of this virus may have important implications not only for pathogenesis and prevention (25), but also for therapy of HCV infection. Like most RNA viruses, HCV circulates in vivo as a complex population of different but closely related viral variants, commonly referred to as a quasispecies (26). Owing to the large reservoir of biologically different genetic variants provided by the quasispecies (27), viral strains that are resistant to IFN may continuously emerge, posing a major challenge for the development of effective therapeutic measures (28, 29). Studies of the HCV quasispecies and its relationship to the outcome of antiviral therapy have been based mostly on PCR–single-stranded conformation polymorphism analysis (30–35), a method that does not permit tracking of individual viral variants present in the quasispecies nor a precise measurement of the genetic distance among such variants (genetic diversity). In contrast, studies using sequence analysis, which allows for precise fingerprinting of variants, have been limited because of the laborious nature of this technique (36–38).
We investigated the composition and molecular evolution of the HCV quasispecies during the course of IFN therapy by tracking individual viral variants in patients with chronic hepatitis C who exhibited different patterns of response. Our main objective was to determine whether the evolution of the viral quasispecies provides biological clues for understanding and predicting the different patterns of treatment response.
Materials and Methods
Patients.
A total of 23 patients with chronic hepatitis C who had been treated with IFN monotherapy between 1994 and 1997 at the Department of Internal Medicine of the University of Cagliari were included in this study. Eight women and 15 men (mean age ± SD, 43.35 ± 2.7, range 25 to 58 years) were selected from a larger number of patients treated with IFN because they represented clear cases of the four different patterns of response to IFN therapy, defined in detail as supporting information on the PNAS web site, www.pnas.org. Of the 23 patients, five had a sustained response, five relapsed after termination of therapy, four had a breakthrough during therapy, five failed to respond, and four untreated patients served as controls. At the time of enrollment the HCV genotype had not been determined and thus the selection of the patients was based only on the different biochemical and virological patterns of response to therapy. All patients had an elevated serum alanine aminotransferase level for at least 6 months before treatment and all had histologically proven chronic hepatitis; 20 of 23 patients (87%) had chronic hepatitis (mild in 11 and moderate in nine); whereas three (13%) had active cirrhosis. There were no significant differences in baseline histology or alanine aminotransferase level (mean ± SD, 147.6 ± 68.3) among the different patient groups. All patients were positive for anti-HCV antibodies by third-generation ELISA assay and for serum HCV RNA as measured by a nested reverse transcription–PCR (39) using two sets of primers from the 5′ noncoding region (40). All were negative for hepatitis B surface antigen and antibodies to HIV type 1 (HIV-1). Patients received IFN-α-2a at a dose of 6 million units, three times a week for 6 months and were followed for at least 3 years after completion of therapy. The protocol was approved by the university's research committee, and all patients provided informed consent. During the treatment period, the patients were seen each week for the first 2 weeks, then biweekly through week 12, and then at 4-week intervals. During the follow-up period, the patients were seen during the first and third months and then every 3 months.
Design of the Study.
The number of viral variants, the genetic distance among the different variants (genetic diversity), the evolution of HCV quasispecies, and the level of viral replication were studied in serial serum samples obtained at different time points during IFN therapy and also at different time points from untreated controls. The analysis of the HCV quasispecies was performed by examining viral sequences spanning the hypervariable region 1 (HVR1) (41) and flanking sequences from the envelope genes [envelope glycoprotein 1 (E1) and E2]. For each patient, we studied one sample obtained at baseline and one within the first 2 weeks of treatment. A later sample was analyzed in all patients (except the long-term responders who cleared the virus) according to the response pattern. In patients who relapsed after therapy, testing was performed on a sample obtained within 1 month after the cessation of therapy. In patients who relapsed during therapy, testing was performed at the time of breakthrough, usually 12 weeks after the initiation of therapy. In nonresponders, sampling was also at 12 weeks after initiation of therapy and in untreated controls at 24 weeks after enrollment.
Test for HCV RNA and Genotype.
Total RNA was extracted from 100 μl of serum and reverse-transcribed in a volume of 20 μl, and the resulting cDNA was amplified by using sets of primers from the E1 and E2 genes, including the HVR1 (40). The sensitivity, specificity, and details of our nested PCR technique have been reported (39). To reduce the risk of contamination, appropriate precautions were taken (42). The level of serum HCV RNA was measured by a commercial assay (Cobas Amplicor HCV Monitor 2.0, Roche Diagnostics). The HCV genotype was determined by sequence analysis of part of the E1 gene (43).
Molecular Cloning and Sequencing.
The PCR products amplified from the E1/E2 region were purified by using Qiaex (Qiagen, Valencia, CA), cloned into pGEM-T vector systems (Promega), and transformed into Escherichia coli strain J109. For sequencing, plasmid DNA was extracted with the Qiaprep Miniprep (Qiagen) according to the manufacturer's recommendations. The double-stranded plasmid DNA was sequenced with the Applied Biosystems model 373 automated DNA sequencer by a modified Sanger method. A total of 1,987 molecular clones, each 558 nt in length, were sequenced.
Data Analysis.
To ensure integrity of the sequence data, appropriate precautions were taken as recommended (44). Genetic diversity was calculated by analysis of predicted amino acid sequences, 176 aa in length, amplified from the E1 and E2 genes of the HCV genome, including the 31 aa of HVR1. The genetic diversity was calculated by the Hamming distance, which is defined as the number of amino acid differences between two sequences (45, 46). The mean Hamming distance, which is the average of the values taken for all sequence pairs derived from a single sample, was separately calculated within the HVR1 (31 aa) and on the entire sequence outside the HVR1 (145 aa). Analysis of the genetic diversity and number of viral variants was performed after exclusion of defective or unreadable sequences, within the HVR1 on 1,790 sequences (mean of 27.9 molecular clones for each sample) and outside the HVR1 on 1240 clones (mean 19.7 molecular clones for each sample). More details on the defective sequences are published as supporting information. The average number of synonymous (silent) nucleotide substitutions per synonymous site and the number of nonsynonymous (amino acid replacement) nucleotide substitutions per nonsynonymous site relative to the ancestral consensus sequence were calculated (47) for each time point within a single patient with the program mega (48). Sequences obtained from each time point were compared with the consensus (reference) sequence of the first time point. The phylogenetic trees were constructed with the neighbor program in the phylip package (49).
Statistical Analysis.
The results are expressed as the mean ± SE. A one-way ANOVA was performed before comparing data of different groups by t test. Changes in the levels of viremia were evaluated by nonparametric Wilcoxon test for paired samples because of the non-normal distribution of data. Frequency variables were compared by a Fisher's exact test. In all tests, a P value less than 0.05 was considered statistically significant.
Results
The five groups of patients were similar with respect to demographic and clinical characteristics, except that 65% of the total population was male and four of the five long-term responders were females. Before treatment, the genetic diversity and the number of viral variants within the HVR1 did not differ significantly among the patient groups (Table 1). However, at baseline, patients with a complete response who later relapsed exhibited the most heterogeneous viral population, with the highest genetic distance among the different viral strains. Similarly, the level of viremia did not differ significantly among the patient groups, although it was slightly higher in nonresponders and controls. Consistent with the geographic origin of the 23 patients, the vast majority, 16 (69.5%) of HCV infections were genotype 1a or 1b (Table 1). One (4.3%) was genotype 2c, and six (26%) were genotype 3a. All nonresponders were infected with genotype 1b, whereas only one of the five patients who exhibited a sustained long-term complete response was infected with genotype 1b (Table 1).
Table 1.
Genetic diversity, number of viral strains, viral genotypes, and levels of serum HCV RNA at baseline in patients with chronic hepatitis C according to their response to IFN-α therapy and in untreated controls
| Sustained response (n = 5) | Relapse (n = 5) | Breakthrough (n = 4) | Nonresponse (n = 5) | No treatment (n = 4) | |
|---|---|---|---|---|---|
| Genetic diversity | |||||
| HVR1 | 8.32 ± 1.98 | 18.60 ± 5.72 | 10.33 ± 3.13 | 10.16 ± 4.40 | 7.50 ± 2.73 |
| E1/E2 outside HVR1 | 1.90 ± 0.18 | 2.30 ± 0.49 | 2.08 ± 0.24 | 2.24 ± 0.49 | 2.52 ± 0.44 |
| No. of viral strains | |||||
| HVR1 | 7.20 ± 0.80 | 11.40 ± 1.75 | 11.50 ± 2.96 | 11.40 ± 2.36 | 9.50 ± 0.87 |
| E1/E2 outside HVR1 | 2.72 ± 0.24 | 3.08 ± 0.33 | 2.97 ± 0.65 | 2.60 ± 0.44 | 3.72 ± 0.50 |
| HCV genotype | |||||
| 1 | 1* | 4 | 3 | 5* | 3 |
| Non-1 | 4 | 1 | 1 | 0 | 1 |
| Serum HCV RNA | |||||
| No. of copies × 105/ml | 8.60 ± 2.91 | 7.86 ± 1.91 | 10.68 ± 4.51 | 12.84 ± 3.50 | 14.35 ± 3.07 |
The analysis of the genetic diversity measured by mean Hamming distance (45) and of the number of viral strains was based on 649 sequences within the HVR1 and on 447 sequences outside the HVR1. The data represent mean ± SEM.
, P = 0.024 for the comparison with the nonresponse group, by Fisher's exact test. None of the other differences were statistically significant.
Patients with a sustained response had a significant decrease both in genetic diversity and number of viral strains (Table 2). These changes were associated with a significant reduction in the levels of HCV viremia. In most patients with a sustained response, the genetic diversity markedly decreased by week 2 (Fig. 1A), leading to a highly homogeneous viral population before the subsequent clearance of HCV viremia, which occurred in all cases within 4 weeks after starting therapy. In contrast, patients who had a complete response followed by relapse showed a disparity between the evolution of the HCV quasispecies and the pattern of viremia at week 2 of therapy. Although this group had a significant decrease in the level of viremia, there was only a slight decrease in genetic diversity and the number of viral strains by week 2 of therapy (Table 2). At the time of relapse, there was reactivation in HCV viremia, which returned to baseline levels, and a further, but not statistically significant, decrease in genetic diversity. Similarly, in patients who had a breakthrough during therapy, HCV RNA levels initially declined in parallel with a reduction in genetic diversity and the number of viral strains, although the extent of such changes did not reach statistical significance (Table 2). These parameters returned to baseline values at the time of breakthrough. In nonresponders, although the level of viremia was almost unchanged, there was a reduction in genetic diversity and in the number of viral strains at week 2 that continued through week 12 of therapy. Despite similar mean values, pairwise comparison showed a significant reduction in both parameters, caused by a more consistent trend in all patients (Table 2). Such a discrepancy suggests that in nonresponders there is persistence of a restricted number of viral strains that have a better fitness for survival during IFN treatment. In untreated controls, there were minimal changes in viral load, number of variants, and genetic diversity over the course of 24 weeks (Table 2).
Table 2.
Changes in genetic diversity, number of viral strains, and levels of serum HCV RNA in patients with chronic hepatitis C according to their response to IFN-α therapy and in untreated controls
| Patient group | No. of patients | Time points | Interval mean, weeks | Change in genetic
diversity*
|
Change in the number of viral
strains
|
Change in serum HCV RNA ×103 copies/ml | ||
|---|---|---|---|---|---|---|---|---|
| HVR1 | E1/E2 outside HVR1 | HVR1 | E1/E2 outside HVR1 | |||||
| Sustained response | 5 | a vs. b | 1.4 ± 0.2 | −5.82 ± 1.46† | −0.74 ± 0.23 | −3.60 ± 2.51‡ | 0.43 ± 0.98 | −859.25 ± 291.26§ |
| Relapse | 5 | a vs. b | 1.8 ± 0.2 | −3.54 ± 1.54 | −0.20 ± 0.19 | −3.60 ± 1.69 | −0.20 ± 0.19 | −770.36 ± 182.69¶ |
| a vs. c | 29.6 ± 1.0 | −7.90 ± 4.42 | −0.92 ± 0.46 | −1.20 ± 2.04 | −0.92 ± 0.46 | −95.10 ± 125.13 | ||
| Breakthrough | 4 | a vs. b | 2.0 ± 0.0 | −4.90 ± 4.03 | −0.93 ± 0.57 | −6.00 ± 3.49 | −1.03 ± 0.54 | −996.81 ± 481.94 |
| a vs. c | 12.0 ± 0.0 | 3.35 ± 5.40 | 0.45 ± 0.98 | −0.25 ± 1.03 | −0.25 ± 0.87 | −282.78 ± 338.80 | ||
| Nonresponse | 5 | a vs. b | 2.0 ± 0.0 | −3.72 ± 1.39 | −0.64 ± 0.40 | −4.60 ± 1.86 | −0.44 ± 0.49 | −95.11 ± 96.50 |
| a vs. c | 12.0 ± 0.0 | −1.38 ± 0.16‖ | −0.16 ± 0.43 | −4.60 ± 1.33** | 0.20 ± 0.41 | −135.79 ± 114.56 | ||
| No treatment | 4 | a vs. b | 2.0 ± 0.0 | −0.03 ± 0.17 | 0.38 ± 0.33 | −1.00 ± 0.91 | −0.03 ± 0.87 | −81.02 ± 312.36 |
| a vs. c | 27.8 ± 1.0 | 0.30 ± 1.51 | 0.45 ± 0.24 | −0.25 ± 1.11 | 1.03 ± 0.61 | −184.80 ± 300.48 | ||
, P = 0.017 for the change between time points a and b, by a paired t test.
, P = 0.033 for the change between time points a and b, by a paired t test.
, P = 0.043 for the change between time points a and b, by nonparametric Wilcoxon test for paired samples.
, P = 0.043 for the change of time points a and b, by nonparametric Wilcoxon test for paired samples.
, P = 0.009 for the change between time points a and c, by a paired t test.
, P = 0.026 for the change between time points a and c, by a paired t test.
Genetic diversity was measured by mean Hamming distance (45). The data represent mean ± SEM. Negative values indicate a reduction in genetic diversity, number of viral strains, and levels of serum HCV RNA; positive values indicate an increase in these values. The first time point (a) corresponds to the baseline, the second (b) to the first or second week of therapy and the third (c) to the time of relapse (mean 29.6 weeks), or breakthrough (week 12), or a late sample in nonresponders (week 12) or in untreated controls (mean 27.8 weeks).
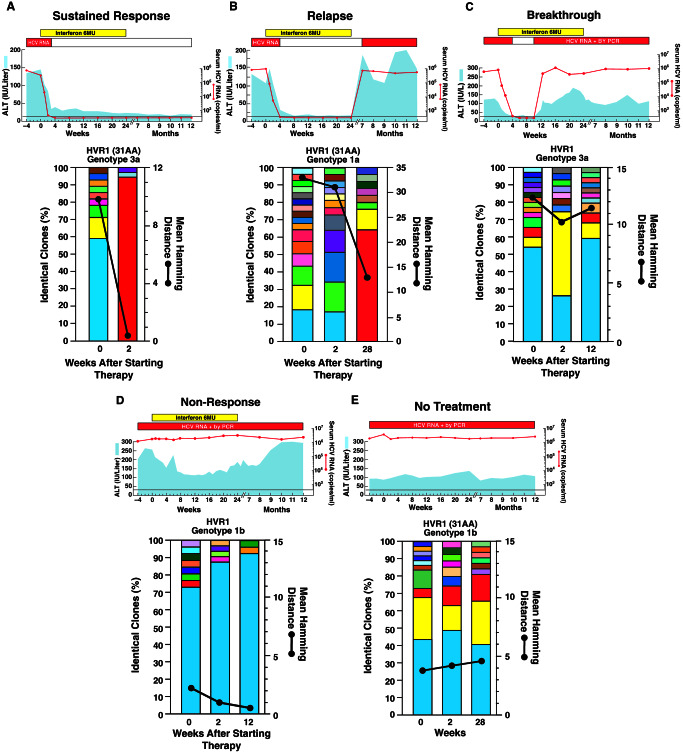
Figure 1.
Clinical course and evolution of the HCV quasispecies within the HVR1 in representative patients according to the pattern of response to IFN therapy (A–D) and in untreated controls (E). (Upper) The clinical course. The light blue areas indicate the values for alanine aminotransferase. The red horizontal bars indicate positive assays for serum HCV RNA by PCR. The red lines indicate the titer of serum HCV RNA as measured by the monitor assay. The yellow horizontal bars indicate the duration and dosage of IFN treatment. (Lower) The number of viral strains and the genetic diversity within the 31 aa of the HVR1. The vertical bars indicate the number and the proportion of viral variants within each sample. Within the vertical bars, each variant is identified by a different color. The dominant viral variant found in each patient at the first time point is indicated in turquoise; other variants are indicated by additional colors. The same color indicates identity between viral variants detected at different time points, but not between different patients. The genetic diversity (black line) was calculated by mean Hamming distance (45) from the predicted amino acid sequences obtained from each sample.
When we performed the analysis on the 145 predicted aa outside the HVR1, the genetic diversity and the number of viral strains at baseline were consistently lower than within the HVR1 in all patient groups (Table 1). Moreover, these parameters outside the HVR1 did not show any change over time (Table 2). Therefore, the patterns observed in patients with different responses to IFN therapy were essentially caused by genetic variation within the HVR1.
Next, we tracked individual viral variants over time by examining HVR1 sequences in patients with different patterns of response to therapy. This region was selected because it is the most variable region of the HCV genome (41) and it has been shown to contain a neutralization domain (50, 51). Thus it might be an important target for monitoring the immunologically mediated antiviral effects induced by IFN. In sustained responders, the analysis conducted at week 2 showed a remarkable loss of most of the strains present at baseline, in parallel with the dramatic decrease in the level of HCV viremia, as depicted in a typical patient (Fig. 1A). In addition, all of the residual strains were present at very low absolute copy number, compared with baseline levels, and in most of the patients a single strain represented on average about 90% of the entire viral population. The dominant strain at baseline disappeared in three patients, while it was still present in the remaining two, although in only one was it dominant. In contrast, the dominant strain persisted in all of the nonresponders, with little, if any, change in the absolute number of viral copies (typical patient: Fig. 1D). This finding, together with a significant reduction in the genetic diversity and the number of viral strains at week 12, is consistent with a higher fitness of the pre-existing dominant strain during therapy. A similar pattern was observed in patients who experienced a breakthrough during therapy (typical patient: Fig. 1C). The dominant strain persisted in three of the four patients at week 2, and in all patients remained the dominant strain at the time of breakthrough.
A more heterogeneous pattern was seen in patients who relapsed after the termination of therapy (Fig. 1B). A distinctive feature of all but one patient with relapse was a high degree of viral diversity at baseline with no single strain representing more than 25% of the total population. At relapse, two patterns were observed. In three cases, there was emergence of a new strain that now showed clear dominance (Fig. 1B) and in two cases an apparently minor population emerged as the distinctly dominant strain.
Untreated controls showed a pattern very similar to that seen in nonresponders in that the dominant strain persisted over time in all patients. A common feature of both untreated and treated patients, regardless of the different patterns of response to therapy, was the continuous replacement of minor variants (each usually represented by a single clone), with new strains during the entire observation period (examples: Fig. 1 A–E). Thus, the major difference among the patterns of response to treatment was the behavior of the dominant strain, which persisted and remained quantitatively predominant in nonresponders, patients with breakthrough, and controls, whereas it disappeared or lost its dominance in almost all patients with a sustained response. In patients with relapse there was no dominant strain at baseline, but an unequivocally dominant strain at relapse.
Phylogenetic analysis of all of the HVR1 amino acid sequences revealed distinct clusters of unique viral sequences for each subject, indicating that the results were not caused by PCR contamination. Phylogenetic reconstructions showed different patterns according to the response to therapy. In most patients with a sustained response, the administration of IFN resulted, by week 2, in the emergence of strains that were distinct from the baseline sequences, clustering as a distinct monophyletic population (typical example: Fig. 2A, which is published as supporting information on the PNAS web site). A tendency to form clusters over time was also seen in most patients with the relapsing pattern. In these patients, however, clustering of the sequences usually occurred at the time of relapse (see Fig. 2B). In contrast, in nonresponders, patients who had a breakthrough, and untreated patients, there was no tendency to form clusters with sampling time. The pattern was characterized by an intermingling of sequences from different time points (typical example: see Fig. 2 C–E).
The accumulation rates of synonymous relative to nonsynonymous nucleotide substitutions revealed a difference in virus evolution within and outside the HVR1 (Table 3, which is published as supporting information on the PNAS web site). In all groups of patients, the rate of nonsynonymous substitutions within the HVR1 was generally higher than that of synonymous substitutions, suggesting a positive selection. Treatment with IFN did not modify this trend, except in patients with a sustained response, in whom synonymous substitutions prevailed at week 2 of therapy. In contrast, the substitutions outside the HVR1 were mainly synonymous in all groups of patients, suggesting that this region evolved under purifying selection (see Table 3).
Discussion
Our study provided evidence that the number of viral strains and the genetic diversity before treatment did not correlate with treatment outcome. Consistent with these observations, phylogenetic analysis of viral sequences obtained from all patients before treatment failed to show any clustering associated with a specific pattern of response. In contrast, within the limitation of our data set, analysis of the early evolution of the viral quasispecies yielded important prognostic information. In patients who exhibited a sustained therapeutic response, we documented a significant decrease in the number of viral strains, genetic diversity, and levels of viremia within 2 weeks of initiating therapy. In contrast, in patients who relapsed after termination of therapy, there were no significant changes in the HCV quasispecies, despite a significant reduction in the level of HCV RNA replication, suggesting that important changes in the HCV quasispecies, but not in viremia, early after starting therapy may differentiate patients who will eradicate the virus from those who will relapse shortly after cessation of therapy. A dramatic reduction in genetic diversity leading to an increasingly homogeneous viral population was a consistent feature associated with viral clearance in sustained responders and was independent of HCV genotype. A strikingly similar trend of decreasing viral diversity was recently documented just before viral clearance in acute resolving hepatitis, whereas an increase in viral diversity was found to correlate with acute hepatitis that progressed to chronicity (52). Although the immunological correlates of spontaneous viral clearance are still undefined (53), it has been suggested that a reduction in genetic diversity in the envelope genes, and especially in the HVR1 of the E2 gene, a neutralization domain, is likely to be the result of a more successful and balanced cellular and humoral immune response (54). Phylogenetic analysis showed a distinct clustering of viral sequences leading to a monophyletic population of variants in sustained responders, suggesting that in these patients there is a shift in HCV quasispecies shortly after initiating therapy, likely as the result of the rapid elimination of IFN-sensitive HCV strains. In contrast, the lack of progressive shift in the viral population seen in nonresponders and those who had a breakthrough suggests a relative evolutionary stasis of the HCV quasispecies. Strikingly, untreated controls showed a pattern most similar to that observed in nonresponders with an intermingling of sequences over time.
The molecular mechanisms underlying treatment failure in chronic HCV infection are still unknown. Our observation that in nonresponders the initial dominant strain persists without significant changes in viral replication levels argues in favor of the hypothesis that inherently IFN-resistant HCV variants are present in these patients before the initiation of therapy. Although a region within the nonstructural protein 5A, named the IFN sensitivity-determining region (11), and a highly conserved stretch of 12 aa in the E2 protein (20) have been suggested as potential determinants of IFN resistance, the evidence for this is inconsistent (25). In this respect and in accordance with several previous studies (14–19, 21–24), we failed to observe any correlation between mutations in these two regions and outcome of IFN therapy (P.F., unpublished data). The mechanism of treatment failure in patients who relapsed during or shortly after therapy is more difficult to interpret. In patients who had a breakthrough, the reappearance of the original dominant strain suggests either an intermediate grade of sensitivity to IFN or, alternatively, transient viral suppression by IFN-modulated immune-mediated mechanisms. In contrast, the emergence of new viral strains in patients who had a relapse suggests that most of the baseline variants were sensitive to IFN. The origin of these new strains is unclear. They could represent new mutants, pre-existing minor variants present at levels below the threshold for detection, or pre-existing variants derived from viral reservoirs that were less accessible to IFN. Regardless of the mechanism, it is surprising that an RNA virus that apparently does not induce latent infection can persist for weeks to months, in both breakthrough and relapse patients, in the face of a seemingly complete suppression of viremia. The most plausible hypothesis is that, despite the disappearance of viremia, very low levels of viral replication continue to occur, most likely in the liver, providing a persistent reservoir for virus reactivation after release from the suppressive effects of IFN.
In conclusion, there are parallels between the spontaneous resolution of acute HCV infection and the response to IFN in chronically infected patients. The early evolution of the HCV quasispecies defines patients who will have a sustained biochemical and virologic resolution of infection. In contrast, measurement of HCV RNA level alone does not distinguish complete responders from those who will subsequently experience a breakthrough or relapse. Thus, the study of the HCV quasispecies, although technically complex, may provide prognostic information as early as 2 weeks after starting therapy. In addition, the study of the HCV quasispecies opens new perspectives for elucidating the mechanisms of treatment failure in chronic hepatitis C. The persistence of variants present before treatment in patients who fail to respond or who experience a breakthrough during therapy strongly suggests the existence of viral strains with inherent resistance to IFN. The genetic basis for this resistance is presently unknown. Further elucidation of the mechanisms of HCV resistance will depend on the development of reliable and sensitive replication models. In the interim much can be learned from the study of the HCV quasispecies early after the initiation of therapy.
Supplementary Material
Acknowledgments
We thank D. Wong, R. Scioscia, and D. Cao for their technical help, P. Lusso for critical reading of the manuscript, and K. Prims and H. Newman for editorial assistance. This study was supported in part by a grant from the Regione Autonoma della Sardegna and from COFIN 2000, Italy.
Abbreviations
- HCV
hepatitis C virus
- E1
envelope glycoprotein 1
- E2
envelope glycoprotein 2
- HVR1
hypervariable region 1
References
- 1.Alter H J, Seeff L B. Semin Liver Dis. 2000;20:17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 2.Houghton M. Curr Top Microbiol Immunol. 2000;242:327–329. doi: 10.1007/978-3-642-59605-6_15. [DOI] [PubMed] [Google Scholar]
- 3.Poynard T, Marcellin P, Lee S S, Niederau C, Minuk G S, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, et al. Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 5.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, et al. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 6.Hoofnagle J H. Adv Intern Med. 1994;39:241–275. [PubMed] [Google Scholar]
- 7.Davis G L, Lau J Y. Hepatology. 1997;26:122S–127S. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 8.Pawlotsky J M. Hepatology. 2000;32:889–896. doi: 10.1053/jhep.2000.19150. [DOI] [PubMed] [Google Scholar]
- 9.Tan S L, Katze M G. Virology. 2001;284:1–12. doi: 10.1006/viro.2001.0885. [DOI] [PubMed] [Google Scholar]
- 10.Taylor D R. Hepatology. 2001;33:1547–1549. doi: 10.1053/jhep.2001.25447. [DOI] [PubMed] [Google Scholar]
- 11.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. J Clin Invest. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 13.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, et al. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 14.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J P, Lunel F, Stuyver L, et al. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 15.Zeuzem S, Lee J H, Roth W K. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 16.Hofgärtner W T, Polyak S J, Sullivan D G, Carithers R L, Jr, Gretch D R. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 17.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 18.Chung R T, Monto A, Dienstag J L, Kaplan L M. J Med Virol. 1999;58:353–358. [PubMed] [Google Scholar]
- 19.Nousbaum J, Polyak S J, Ray S C, Sullivan D G, Larson A M, Carithers R L., Jr J Virol. 2000;74:9028–9038. doi: 10.1128/jvi.74.19.9028-9038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M. Science. 1999;285:107–110. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 21.Abid K, Quadri R, Negro F. Science. 2000;287:1555. doi: 10.1126/science.287.5458.1555a. [DOI] [PubMed] [Google Scholar]
- 22.Chayama K, Suzuki F, Tsubota A, Kobayashi M, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Takahashi N, et al. Hepatology. 2000;32:1138–1144. doi: 10.1053/jhep.2000.19364. [DOI] [PubMed] [Google Scholar]
- 23.Sarrazin C, Kornetzky I, Ruster B, Lee J H, Kronenberger B, Roth W K, Zeuzem S. Hepatology. 2000;31:1360–1370. doi: 10.1053/jhep.2000.7987. [DOI] [PubMed] [Google Scholar]
- 24.Berg T, Mas Marques A, Hohne M, Wiedenmann B, Hopf U, Schreier E. Hepatology. 2000;32:1386–1395. doi: 10.1053/jhep.2000.20527. [DOI] [PubMed] [Google Scholar]
- 25.Farci P, Purcell R H. Semin Liver Dis. 2000;20:103–126. [PubMed] [Google Scholar]
- 26.Martell M, Esteban J L, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eigen M, Biebricher C K. In: RNA Genetics. Domingo E, Holland J J, Ahlquist P, editors. Boca Raton, FL: CRC; 1988. pp. 211–245. [Google Scholar]
- 28.Coffin J M. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 29.Domingo E. J Viral Hepat. 1996;2:247–261. [Google Scholar]
- 30.Moribe T, Hayashi N, Kanazawa Y, Mita E, Fusamoto H, Negi M, Kaneshige T, Igimi H, Kamada T, Uchida K. Gastroenterology. 1995;108:789–795. doi: 10.1016/0016-5085(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 31.Koizumi K, Enomoto N, Kurosaki M, Murakami T, Izumi N, Marumo F, Sato C. Hepatology. 1995;22:30–35. [PubMed] [Google Scholar]
- 32.Gonzales-Peralta R P, Qian K, She J Y, Davis G L, Ohno T, Mizokami M, Lau J Y. J Med Virol. 1996;49:242–247. doi: 10.1002/(SICI)1096-9071(199607)49:3<242::AID-JMV14>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 33.Toyoda H, Kumada T, Nakano S, Takeda I, Sugiyama K, Osada T, Kirivama S, Sone Y, Kinoshita M, Hadama T. J Hepatol. 1997;26:6–13. doi: 10.1016/s0168-8278(97)80002-5. [DOI] [PubMed] [Google Scholar]
- 34.Le Guen B, Squadrito G, Nalpas B, Berthelot P, Pol S, Brechot C. Hepatology. 1997;25:1250–1254. doi: 10.1002/hep.510250531. [DOI] [PubMed] [Google Scholar]
- 35.Pawlotsky J M, Germanidis G, Frainais P O, Bouvier M, Soulier A, Pellerin M, Dhumeaux D. J Virol. 1999;73:6490–6499. doi: 10.1128/jvi.73.8.6490-6499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. Hepatology. 1992;16:619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- 37.Kanazawa Y, Hayashi N, Mita E, Tiancheng L, Hagiwara H, Kasahara A, Fusamoto H, Kamada T. Hepatology. 1994;20:1121–1130. [PubMed] [Google Scholar]
- 38.Sandres K, Dubois M, Pasquier C, Payen J L, Alric L, Duffaut M, Vinel J P, Pascal J P, Puel J, Izopet J. J Virol. 2000;74:661–668. doi: 10.1128/jvi.74.2.661-668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farci P, Alter H J, Wong D, Miller R H, Shih J W, Jett B, Purcell R H. N Engl J Med. 1991;325:98–104. doi: 10.1056/NEJM199107113250205. [DOI] [PubMed] [Google Scholar]
- 40.Farci P, Alter H J, Govindarajan S, Wong D C, Engle R, Lesniewski R R, Mushahwar I K, Desai S M, Miller R H, Ogata N, et al. Science. 1992;258:135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 41.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, et al. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwok S, Higuchi R. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 43.Bukh J, Purcell R H, Miller R H. Proc Natl Acad Sci USA. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Learn G H, Jr, Korber B T, Foley B, Hahn B H, Wolinsky S M, Mullins J L. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamming R W. Coding and Information Theory. 2nd ed. Englewood Cliffs, NJ: Prentice–Hall; 1986. [Google Scholar]
- 46.Ganeshan S, Dickover R E, Korber B T, Bryson Y J, Wolinsky S M. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nei M, Gojobori T. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis. University Park: Pennsylvania State Univ.; 1993. [Google Scholar]
- 49.Felsenstein J. phylip: Phylogeny Inference Package. Seattle: Univ. of Washington; 1995. [Google Scholar]
- 50.Farci P, Shimoda A, Wong D, Cabezon T, De Gioannis D, Strazzera A, Shimizu Y, Shapiro M, Alter H J, Purcell R H. Proc Natl Acad Sci USA. 1996;93:15394–15399. doi: 10.1073/pnas.93.26.15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H, Yoshikura H. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 52.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder J C, Strazzera A, Chien D Y, Munoz S J, Balestrieri A, et al. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 53.Rehermann B. Semin Liver Dis. 2000;20:127–141. doi: 10.1055/s-2000-9946. [DOI] [PubMed] [Google Scholar]
- 54.Klenerman P, Lechner F, Kantzanou M, Ciurea A, Hengartner H, Zinkernagel R. Science. 2000;289:2003. doi: 10.1126/science.289.5487.2003a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.