Abstract
Wild-type p53 is a tumor-suppressor gene that encodes a short-lived protein that, upon accumulation, induces growth arrest or apoptosis. Accumulation of p53 occurs mainly by posttranslational events that inhibit its proteosomal degradation. We have reported previously that inhibition of NAD(P)H: quinone oxidoreductase 1 (NQO1) activity by dicoumarol induces degradation of p53, indicating that NQO1 plays a role in p53 stabilization. We now have found that wild-type NQO1, but not the inactive polymorphic NQO1, can stabilize endogenous as well as transfected wild-type p53. NQO1-mediated p53 stabilization was especially prominent under induction of oxidative stress. NQO1 also partially inhibited p53 degradation mediated by the human papilloma virus E6 protein, but not when mediated by Mdm-2. Inhibitors of heat shock protein 90 (hsp90), radicicol and geldanamycin, induced degradation of p53 and suppressed p53-induced apoptosis in normal thymocytes and myeloid leukemic cells. Differences in the effectiveness of dicoumarol and hsp90 inhibitors to induce p53 degradation and suppress apoptosis in these cell types indicate that NQO1 and hsp90 stabilize p53 through different mechanisms. Our results indicate that NQO1 has a distinct role in the regulation of p53 stability, especially in response to oxidative stress. The present data on the genetic and pharmacologic regulation of the level of p53 have clinical implications for tumor development and therapy.
Wild-type p53 is a tumor-suppressor gene that is mutated in more than 50% of human cancers (reviewed in refs. 1 and 2). The tumor-suppressing activity of wild-type p53 is a result of its ability to induce growth arrest (1, 2) or apoptosis (3–6). Wild-type p53 is a short-lived protein (7), and its cellular level is controlled by the rate of its degradation in the proteasomes. This degradation is mainly regulated by association with the E3 ubiquitin ligase protein Mdm-2 that ubiquitinates p53 and targets it to the proteasomes (8, 9). After γ-irradiation or other types of stress, p53 and Mdm-2 undergo posttranslational modifications that diminish their association, leading to diminished p53 degradation (reviewed in ref. 2). Mdm-2 expression is induced by wild-type p53, thus creating a negative feedback loop that maintains p53 at low levels (10, 11). Unlike wild-type p53 in normal cells, mutant p53 accumulates in cancer cells because of its inability to induce expression of Mdm-2 (12) and its formation of ternary complexes with Mdm-2 and heat shock protein 90 (hsp90), which prevents mutant p53 degradation (13). After treatment with hsp90 inhibitors, these complexes are disrupted (13), resulting in degradation of the mutant p53 protein (13–16). However, hsp90 inhibitors such as geldanamycin and radicicol were reported not to cause a similar degradation of wild-type p53 in some cancer cells (14–16).
We have shown previously that dicoumarol, an inhibitor of NAD(P)H: quinone oxidoreductase 1 (NQO1), caused degradation of wild-type p53 in various cell types and suppressed its ability to induce apoptosis in normal thymocytes and in myeloid leukemic cells (17). Dicoumarol also caused destabilization of mutant p53 (17). These results indicated that NQO1 and possibly also other oxidoreductases play an important role in the regulation of the stability of wild-type as well as of mutant p53 and that this also has implications for tumor development and therapy. In this context, it is particularly interesting that NQO1 knockout mice (18) and a genetic polymorphism of NQO1 in humans that results in the loss of its oxidoreductase activity (19–22) are associated with increased susceptibility to tumor development. We now have found that wild-type NQO1, but not the polymorphic NQO1, can stabilize endogenous as well as transfected wild-type p53. NQO1 also partially inhibited p53 degradation mediated by the human papilloma virus E6 protein but not when mediated by Mdm-2. Comparison of the effectiveness of the NQO1 and hsp90 inhibitors to induce p53 degradation and suppress p53-mediated apoptosis indicated that NQO1 and hsp90 act through different mechanisms. Our data indicate that NQO1 has a distinct role in the regulation of p53 stability especially in response to oxidative stress.
Materials and Methods
Cells and Cell Culture.
The cell lines used were HCT116 human colon carcinoma cells, HCT116 HA-NQO1 overexpressing cells (17), p53 null HCT116 cells (23), normal thymocytes obtained from 2.5-month-old Balb/C mice, 7-M12 mouse myeloid leukemic cells (24), and M1-t-p53 mouse myeloid leukemic cells that express a temperature-sensitive p53 [Val-135] protein (3). The p53 in M1-t-p53 cells behaves as a tumor-suppressing wild-type p53 at 32°C and as a mutant p53 at 37°C (25). HCT116 cells were grown in DMEM supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin and cultured at 37°C in a humidified incubator with 5.6% CO2. Normal thymocytes and 7-M12 and M1-t-p53 cells were grown in DMEM supplemented with 10% heat-inactivated (56°C, 30 min) horse serum and cultured at 37°C in an incubator with 10% CO2.
Compounds.
Dicoumarol (Sigma) was dissolved in 0.13 M NaOH. Radicicol and geldanamycin (Calbiochem) were dissolved in DMSO.
Plasmids.
The plasmids used were pRc/CMV (cytomegalovirus) human p53 (26), pEFIRES HA-NQO1 (17), pCOC-mouse mdm2 X2 (26), and pRc/CMV-E6 and pCGN-HA-LT (obtained from U. Nudel, Weizmann Institute of Science). To generate the C609T polymorphism in NQO1, site-directed mutagenesis of HA-NQO1 was performed by PCR by using PWO TaqDNA polymerase (Boehringer Mannheim) and the appropriate sense and antisense primers. The wild-type HA-NQO1and the C609T polymorphic HA-NQO1 were cloned into the pSG5 vector, and their sequence was verified by DNA sequencing.
Transfection.
Cells were seeded at 60% confluence in six-well plates 16 h before transfection with the desired plasmids. Transfections were carried out by the calcium phosphate method followed, 7 h posttransfection, by a 10% glycerol shock for 30 s. The exact amount of plasmid used in each experiment is indicated in the corresponding figure legend. Whenever needed, an empty vector was used to maintain a constant amount of 5 μg total DNA in each transfection mix. Cell extracts generally were prepared 24 h after transfection.
Immunoblot Analysis.
Cell extracts and immunoblot analysis were carried out as described (17). The antibodies used were monoclonal anti-human p53 (PAb 1801) (27), monoclonal anti-mouse Mdm-2 (PAb 4B2) (28), monoclonal anti-mouse and human p53 (PAb240), rabbit anti-IκB (Santa Cruz Biotechnology), hamster anti-mouse Bcl-2 (3F11) (PharMingen), and monoclonal anti-actin (AC-40) and anti-hemagglutinin (anti-HA) (Sigma).
Apoptosis and Cell Viability Assays.
Apoptosis in normal thymocytes was induced by γ-irradiation at 4 Gy (Co60 source, 0.63 Gy/min) and in M1-t-p53 cells by culture at 32°C for 23 h. The percentage of apoptotic thymocytes was determined as described (17) on May–Grünwald–Giemsa-stained cytopsin preparations by counting 400 cells at 5 h post-γ-irradiation. Apoptotic M1-t-p53 cells undergo secondary changes including uptake of trypan blue (29). The percentage of viable cells (nonapoptotic and not stained with trypan blue) was determined by counting 400 cells in a hemacytometer.
Results
Stabilization of p53 by NQO1 Depends on Its Enzymatic Activity.
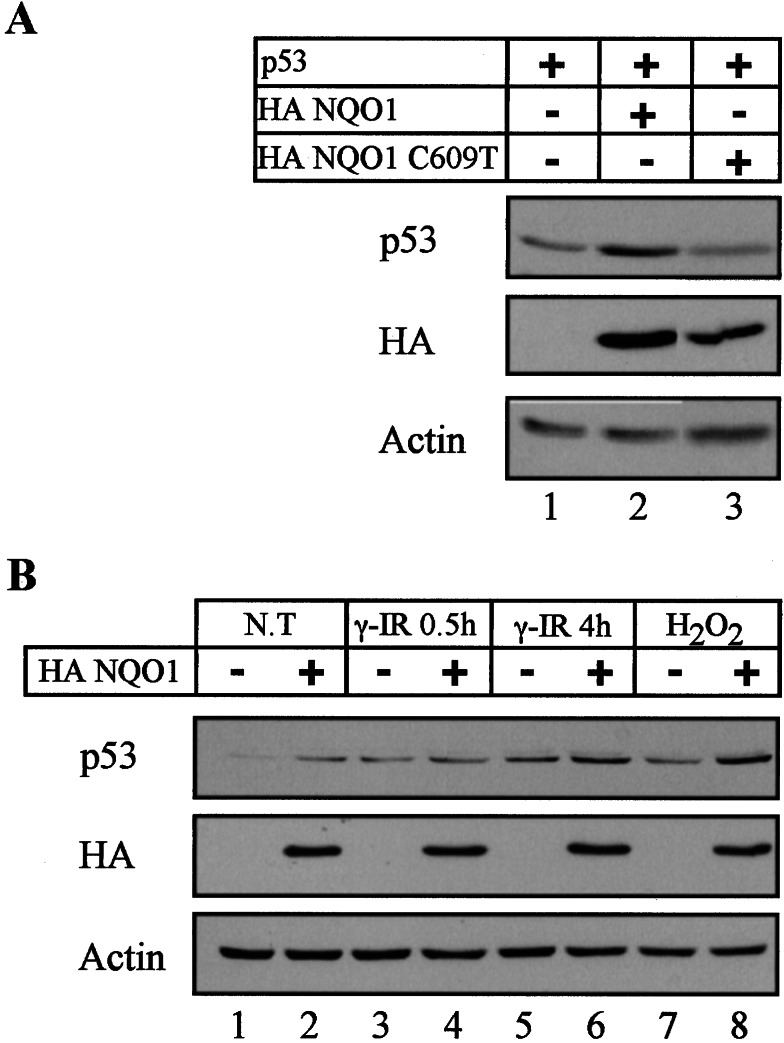
We have shown previously that inhibition of NQO1 activity by dicoumarol induces degradation of wild-type p53 protein in normal thymocytes, myeloid leukemic cells, and HCT116 carcinoma cells but not in HCT116 cells that overexpressed NQO1 (17). To determine directly whether the enzymatic activity of NQO1 is required for p53 stability, we generated a mutant NQO1 expression vector with a C-to-T base pair substitution at position 609 of NQO1 cDNA (C609T). This mutation is a genetic polymorphism in humans that results in a proline-to-serine substitution at residue 187 associated with loss of enzyme activity (30). As expected, cotransfection of p53 null HCT116 cells (23) with wild-type p53 and wild-type NQO1 expression vectors stabilized the level of transfected p53 protein (Fig. 1A, lanes 1 and 2). Unlike the wild-type NQO1, C609T polymorphic NQO1 did not stabilize the cotransfected p53 (Fig. 1A, lanes 1–3). It should be pointed out, however, that the level of C609T NQO1 expressed in the transfected cells was lower than the wild-type NQO1 (Fig. 1B, lanes 2 and 3). Repeated attempts to increase the level of C609T NQO1 expression failed, possibly because the mutant protein is highly unstable (31). The results indicate that stabilization of p53 by NQO1 depends on an intact oxidoreductase activity of the enzyme. An interesting implication of this finding is that individuals with NQO1 C609T polymorphism are likely to suffer from improper regulation of p53 stability.
Figure 1.
NQO1 activity-dependent stabilization of p53 protein. (A) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type p53 with either pSG5 empty vector, 2 μg of pSG5 wild-type HA NQO1, or 3 μg of polymorphic HA C609T NQO1. (B) HCT116 (−) and HCT116 stably expressing HA NQO1 (+) cells were cultured without any treatments (N. T.), γ-irradiated (γ-IR) at 6 Gy, or treated with 100 μM H2O2. Cell extracts were prepared from untreated cells and from cells cultured for a half-hour and 4 h post-γ-irradiation and 6 h after the addition of H2O2. Protein extraction and immunoblot analysis were carried out as described in Materials and Methods by using PAb 1801 monoclonal anti-p53 antibody. The blots then were stripped and reprobed with monoclonal anti-Ha for the detection of HA NQO1 and with anti-actin antibody as a control for equal protein loading in each lane.
Stabilization of p53 Protein by NQO1 Is Especially Prominent under Oxidative Stress.
We next determined the effect of transfected NQO1 on the endogenous p53 level under different physiological conditions. Under normal growth conditions, NQO1 stably transfected HCT116 cells had an elevated level of p53 compared with the parental nontransfected cells (Fig. 1B, lanes 1 and 2). NQO1 transfected cells had about the same level of p53 as the parental cells 0.5 h after γ-irradiation, and there was some increase in p53 at 4 h after γ-irradiation (Fig. 1B, lanes 3–6). The p53 stabilizing effect of NQO1 was more prominent after induction of oxidative stress by H2O2 (Fig. 1B, lanes 7 and 8). These findings suggest that NQO1 interferes with a pathway that leads to p53 degradation, especially under oxidative stress.
NQO1 Does Not Antagonize the Effect of Overexpressed Mdm-2 on p53 Degradation.
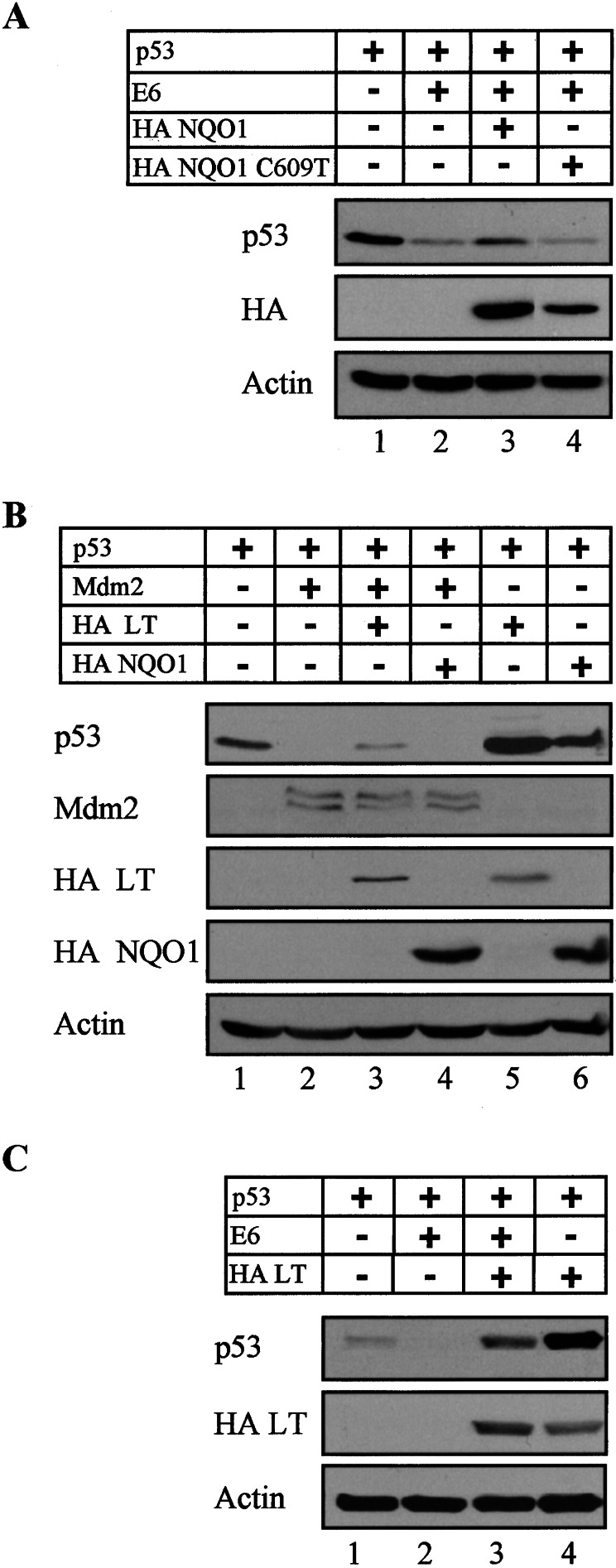
The cellular protein Mdm-2 and the human papilloma virus protein E6 can promote ubiquitination and proteosomal degradation of wild-type p53 whereas the simian virus 40 (SV40) large T antigen (LT) can promote p53 stabilization (reviewed in refs. 1 and 2). In view of the ability of NQO1 to promote p53 stabilization, we determined whether NQO1 can antagonize the effect of overexpressed E6 and Mdm-2 on p53 degradation. P53 null HCT116 cells were transiently transfected with wild-type p53 alone or cotransfected with different combinations of p53, E6, Mdm-2, wild-type NQO1, and the C609T polymorphic NQO1. The results show that E6-mediated p53 degradation could be partially inhibited by cotransfected wild-type NQO1 but not by the C609T polymorphic NQO1 (Fig. 2A). In contrast, although NQO1 stabilized p53 (Fig. 2B, lanes 1 and 6), it did not inhibit the effect of overexpressed Mdm-2 on p53 degradation (Fig. 2B, lanes 2 and 4). The NQO1-mediated p53 stabilization thus is mediated via a pathway that is distinct from that of Mdm-2.
Figure 2.
NQO1 partially antagonizes papillomavirus E6 but not Mdm-2-mediated degradation of p53. (A) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type p53 without and with 500 ng of pRc/CMV-E6, 2 μg of pSG5 wild-type HA NQO1, or 3 μg of polymorphic HA C609T NQO1. (B) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type p53 without or with 300 ng of pCOC-mdm2 X2, 1.5 μg of pCGN-HA-LT, or 2 μg of pSG5 wild-type HA NQO1. (C) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type p53 without or with 800 ng of pRc/CMV-E6 or 1.5 μg of pCGN-HA-LT. Immunoblot analysis was carried out by using PAb 1801 monoclonal anti-p53 antibody. The blots then were stripped and reprobed with monoclonal anti-Mdm-2 and anti-HA for the detection of HA NQO1 and HA LT, and anti-actin antibody as a control for equal protein loading in each lane.
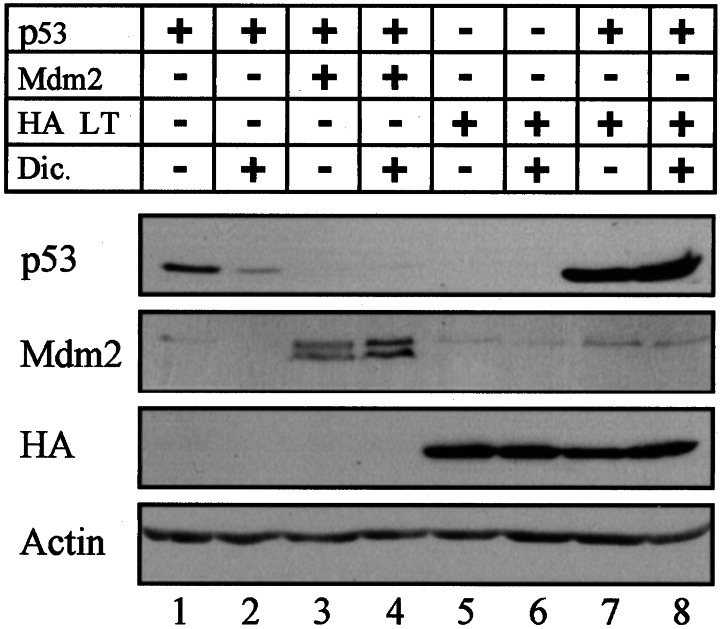
We previously have reported that the NQO1 inhibitor dicoumarol induced p53 degradation in various cell types, but not in COS-1 cells that express LT (17). This indicated that inhibition of NQO1 activity could not promote p53 degradation in cells in which p53 was already stabilized by LT. To test this directly in the same cell type, p53 null HCT116 cells were transiently transfected with different combinations of wild-type p53, Mdm-2, E6, NQO1, and LT. The results indicate that cellular p53 was elevated to higher levels in LT compared with NQO1-transfected cells (Fig. 2B, lanes 1, 5, and 6). Unlike NQO1, LT partially inhibited the ability of Mdm-2 to promote p53 degradation (Fig. 2B, lanes 3 and 4). In addition, LT appeared to be more potent than NQO1 in antagonizing the p53-degradation effect of E6 (Fig. 2 A and C, lanes 2 and 3). Furthermore, as in our previous results with LT-expressing COS-1 cells (17), HCT 116 cells cotransfected with p53 and LT were resistant to dicoumarol-induced p53 degradation (Fig. 3, lanes 7 and 8), whereas cells transfected only with p53 were sensitive to dicoumarol-induced p53 degradation (Fig. 3, lanes 1 and 2). The levels of both Mdm-2 (Fig. 3, lanes 3 and 4) and LT (Fig. 3, lanes 5–8) were not affected by dicoumarol. This indicates that dicoumarol did not affect transfection efficiency or the stability of Mdm-2 and LT. Our data indicate that both LT and NQO1 stabilize p53 and block dicoumarol-mediated p53 degradation (7). However, unlike LT, NQO1 does not affect p53 degradation mediated by overexpressed Mdm-2.
Figure 3.
LT protects p53 protein from degradation induced by dicoumarol. p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type p53 without or with 300 ng of pCOC-mdm2 X2 or 1.5 μg of pCGN-HA-LT. Transfected cells (24 h posttransfection) then were cultured for 5 h without or with 300 μM dicoumarol.
Different Mechanisms for p53 Stabilization by NQO1 and hsp90.
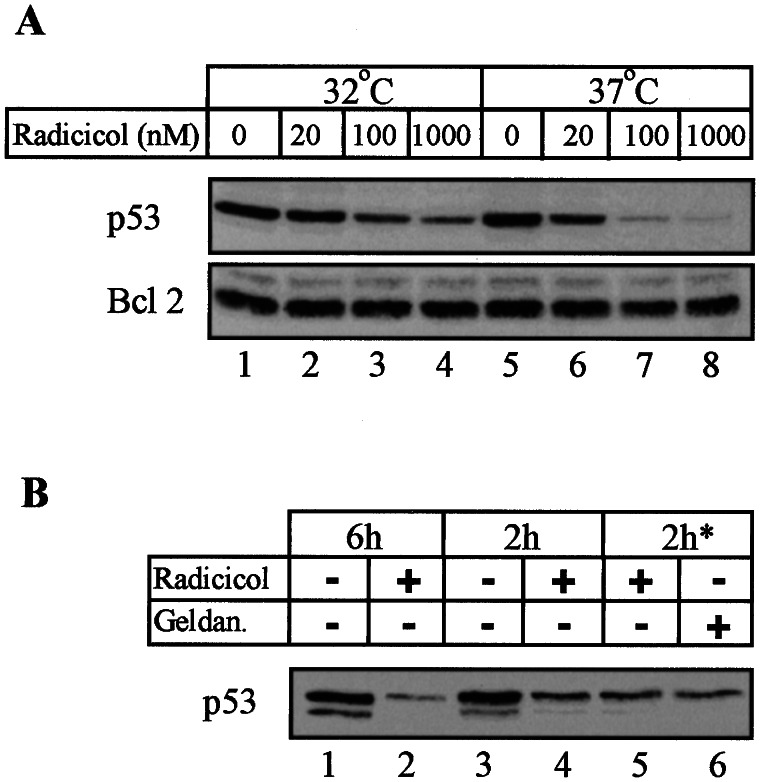
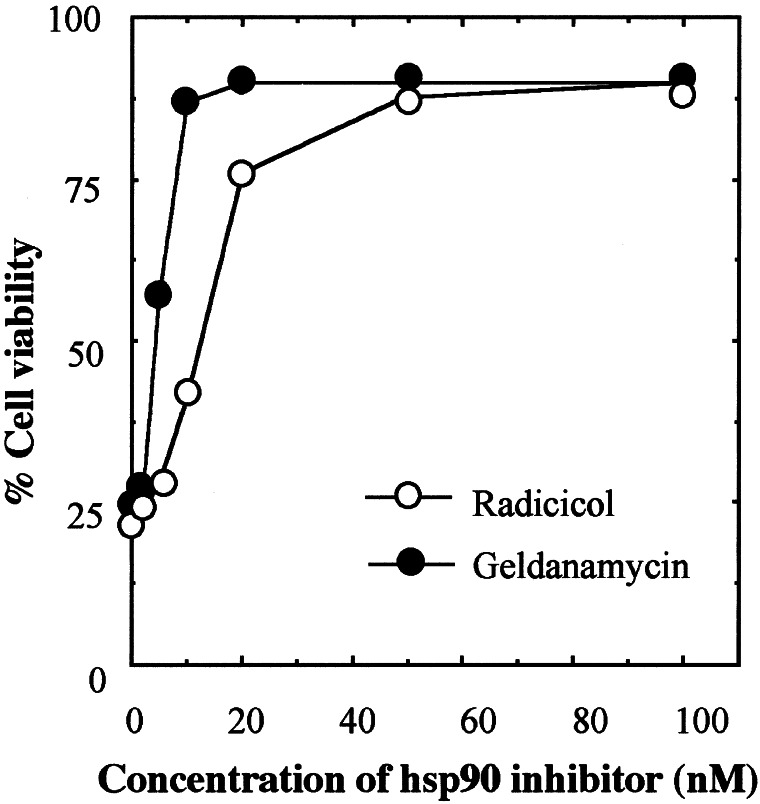
Using different types of tumor cells carrying wild-type or mutant p53, others have reported that hsp90 interacts with mutant p53 but not with wild-type p53 (13, 15, 16, 32) and that hsp90 inhibitors could disrupt this interaction (13, 16, 32) and cause degradation of mutant p53 (13–16, 32). However, work with the ras-transformed rat embryo fibroblast cell line A1, which expresses the temperature-sensitive Val-135 mutant p53, has shown that hsp90 inhibitors also can cause degradation of p53 at 32°C when the protein assumes wild-type conformation (15). We have shown that the same temperature-sensitive Val-135 mutant p53 can be degraded in its mutant and wild-type forms in M1-t-p53 myeloid leukemic cells by the NQO1 inhibitor dicoumarol (17). This raised the possibility that NQO1 and hsp90 inhibitors may induce p53 degradation in certain cell types through a similar mechanism. Using M1-t-p53 leukemic cells, we therefore determined whether hsp90 inhibitors also can cause degradation of p53 in its mutant form at 37°C and its wild-type form at 32°C. The results indicate that a 6-h incubation of M1-t-p53 cells with the hsp90 inhibitor radicicol caused a strong decrease in the level of p53 both in its mutant form (at 37°C) and in its wild-type form (at 32°C) (Fig. 4A). The decrease at 37°C appeared to be stronger than at 32°C. Under the same conditions, there was no change in the level of Bcl-2 (Fig. 4A), indicating that the radicicol-induced decrease in p53 level was not due to a general effect on the stability of all cellular proteins. To ensure that the radicicol-induced decrease in p53 in cells shifted to 32°C was not due to a rapid degradation of mutant p53 before it could assume a wild-type conformation, we preincubated the cells at 32°C for 4 h and then added radicicol. The results showed that even a 2-h incubation with radicicol at 32°C was sufficient to cause a decrease in p53 level, irrespective of whether radicicol was added at the time of shift from 37°C to 32°C or 4 h after the temperature shift (Fig. 4B, lanes 3–5). Another hsp90 inhibitor, geldanamycin, also showed the same decrease in p53 level at 32°C (Fig. 4B, lane 6). Both radicicol and geldanamycin strongly suppressed the ability of the wild-type p53 to induce apoptosis in the M1-t-p53 myeloid leukemic cells cultured at 32°C (Fig. 5). Cell viability at 32°C reached 90 ± 4% in the presence of 100 nM radicicol or geldanamycin (Fig. 5) and, thus, was more effective than dicoumarol, which, at the optimum concentration of 75 μM, increased cell viability only to 52 ± 4% (17).
Figure 4.
Induction of wild-type and mutant p53 degradation by hsp90 inhibitors. (A) M1-t-p53 myeloid leukemic cells were cultured for 6 h at 32°C or 37°C without or with different concentrations of radicicol. (B) Cells were cultured at 32°C for 6 h or 2 h without (−) or with (+) 1 μM radicicol. Cells also were preincubated for 4 h at 32°C and then cultured with 1 μM radicicol or 1 μM geldanamycin (geldan.) for 2 h (2 h*). Immunoblot analysis was carried out by using PAb 240 monoclonal anti-p53 antibody and hamster anti-mouse Bcl-2.
Figure 5.
Suppression of wild-type p53-mediated apoptosis by hsp90 inhibitors. M1-t-p53 cells were cultured at 32°C without or with different concentrations of radicicol or geldanamycin. Cell viability was determined 23 h after culture at 32°C.
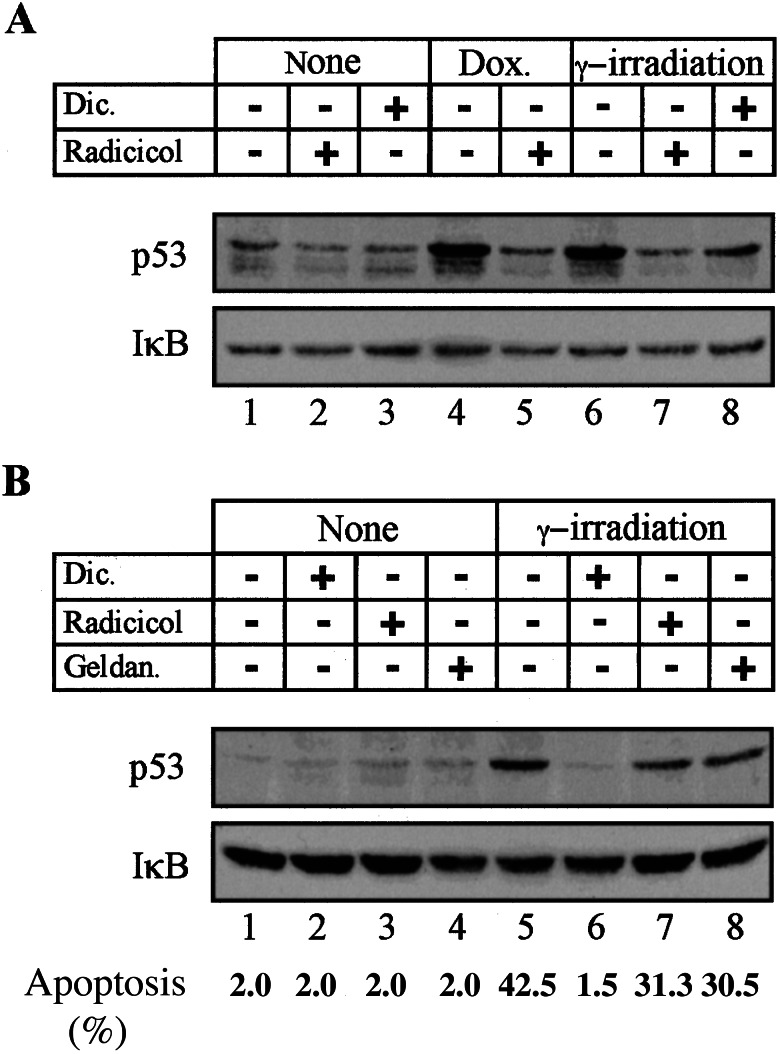
We also have determined the effect of dicoumarol, radicicol, and geldanamycin on p53 level in γ-irradiated or doxorubicin-treated 7-M12 myeloid leukemic cells and in γ-irradiated normal thymocytes. Accumulation of p53 in γ-irradiated or doxorubicin-treated 7-M12 myeloid leukemic cells (29) induced expression of mdm-2 and waf-1 (data not shown). This indicates that p53 in these cells functions as wild-type p53. Analysis of the level of p53 in 7-M12 leukemic cells showed that accumulation of p53 after γ-irradiation or doxorubicin treatment (Fig. 6A, lanes 1, 4, and 6) was inhibited by dicoumarol and radicicol, with radicicol showing a stronger effect (Fig. 6A, lanes 6–8). As shown previously (17), dicoumarol completely inhibited wild-type p53 accumulation in γ-irradiated normal thymocytes (Fig. 6B, lanes 5 and 6). However, radicicol and geldanamycin showed only a weak inhibition of p53 accumulation in thymocytes (Fig. 6B, lanes 5, 7, and 8). Induction of apoptosis in these γ-irradiated thymocytes was also completely inhibited by dicoumarol but only weakly affected by the hsp90 inhibitors (Fig. 6B). These weak effects were not due to a general thymocyte unresponsiveness to hsp90 inhibitors, because both radicicol and geldanamycin strongly suppressed dexamethasone-induced apoptosis in thymocytes (from 42 ± 3% to 5 ± 2% apoptotic cells). The results show that there appear to be cell type-specific differences in the ability of NQO1 and hsp90 inhibitors to decrease wild-type p53 level and apoptosis in normal thymocytes and myeloid leukemic cells. These differences indicate that NQO1 and hsp90 affect intracellular levels of p53 through different mechanisms.
Figure 6.
Degradation of p53 in 7-M12 myeloid leukemic cells and normal thymocytes by dicoumarol and radicicol. (A) 7-M12 cells either were not treated (none), γ-irradiated at 0.4 Gy, or treated with 2.1 μM doxorubicin (Dox.). Cells were cultured for 2 h without (−) or with 100 μM dicoumarol (Dic.) or 5 μM radicicol. (B) Normal thymocytes either were not treated (none) or γ-irradiated at 0.4 Gy and cultured without (−) or with 200 μM dicoumarol (Dic.), 5 μM radicicol, or 5 μM geldanamycin (Geldan.). Cell extracts were prepared after 2 h, and apoptosis was determined after 5 h. Immunoblot analysis was carried out by using PAb 240 monoclonal anti-p53 antibody and rabbit anti-IκB.
Discussion
Wild-type p53 protein is an important regulator of cell fate by its ability to accumulate after DNA damage and other types of stress and to cause growth arrest or apoptosis (reviewed in refs. 1, 2, and 33–35). These properties endow p53 with a tumor-suppressing activity that prevents propagation of cells that were exposed to genotoxic or other types of stress. To prevent death, normal cells therefore must maintain p53 level under tight control. This is achieved mainly by the Mdm-2 protein that associates with p53 and, by virtue of its ubiquitin ligase activity, targets p53 for proteosomal degradation (8, 9). Failure to maintain wild-type p53 at low levels, as occurs in Mdm-2-deficient mice, results in early embryonic lethality that can be prevented by knocking out p53 (36, 37). Posttranslational modifications of p53 and Mdm-2, after γ-irradiation or oxidative stress, result in their diminished association followed by decreased p53 ubiquitination and degradation, thus allowing accumulation of biologically active p53 (reviewed in refs. 2 and 33).
Our present results indicate that overexpressed NQO1 induces p53 accumulation in a manner that cannot be explained by blocking Mdm-2 function. This conclusion is based on the present finding that NQO1 stabilizes p53 under conditions in which the effect of Mdm-2 on p53 is already minimal, such as γ-irradiation or H2O2 treatment. In addition, NQO1 did not block p53 degradation mediated by Mdm-2 overexpression. Mdm-2 is not the only cellular protein that physically interacts with wild-type p53 and regulates its stability. Other proteins include several chaperones and cochaperones, such as hsp40, hsc70, and hsp90, that can stabilize p53 by interfering with the ability of Mdm-2 to target p53 for degradation (13–16, 38). These interactions can be disrupted pharmacologically by the hsp90 inhibitors radicicol and geldanamycin, leading to p53 degradation. Based on the differences in the effectiveness of the NQO1 inhibitor dicoumarol and these hsp90 inhibitors to induce degradation of wild-type p53 and to suppress apoptosis in different cell types, we conclude that NQO1 and hsp90 stabilize p53 through different pathways. This conclusion is supported by the finding that, whereas hsp90 appears to inhibit Mdm-2-mediated degradation of p53 (13), NQO1 expression did not inhibit Mdm-2-mediated p53 degradation. Furthermore, our results show that p53 stabilized by SV40 LT could be degraded by Mdm-2 and E6 but not by the NQO1 inhibitor dicoumarol. The results thus suggest that NQO1 can stabilize p53 by suppressing a distinct pathway of degradation. In analogy to Mdm-2, physical interaction of NQO1 with p53 is an attractive possibility. Studies with WOX1 oxidoreductase has shown that it physically interacts with the proline-rich region of p53, increases the level of p53, and enhances the ability to induce apoptosis (39). It will be interesting to determine whether the same holds for NQO1.
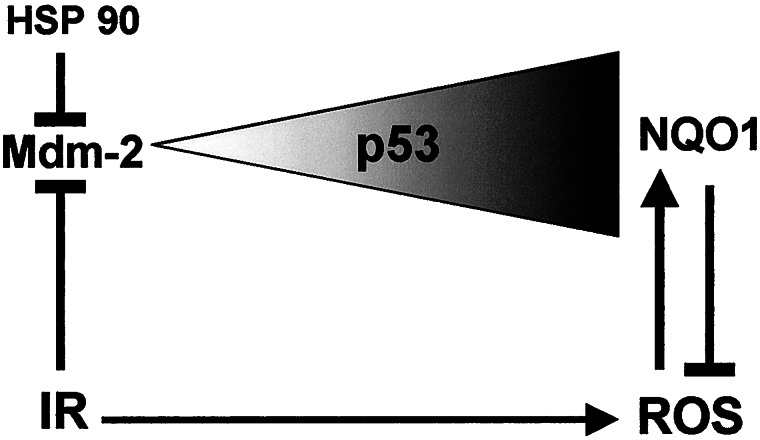
We propose a model whereby p53 level is determined by two opposing effectors, namely, Mdm-2 and NQO1, acting independently of each other (Fig. 7). p53 is stabilized after γ-irradiation or production of reactive oxygen species (ROS), and NQO1-mediated stabilization of p53 is maximal under conditions of increased ROS. ROS is known to induce expression of NQO1, which, in turn, reduces ROS (40). A possible cross-talk between these two pathways is mediated by the production of ROS after γ-irradiation (Fig. 7). In addition, p53 can change the degree of oxidative stress in cells by induction or repression of genes that regulate ROS production (41–45).
Figure 7.
Model of the role of NQO1 in p53 accumulation. The model assumes that the level of p53, depicted as a triangle, is oppositely regulated by Mdm-2 and by NQO1. These pathways function independently, with a possible cross-talk between them mediated by production of ROS after γ-irradiation (IR). ROS increases the level of NQO1, which, in turn, reduces ROS by its oxidoreductase activity in a negative feedback loop. ↓, pathway; ⊥, inhibition of a pathway.
Our results also have biological implications concerning tumor development as well as tumor therapy. It was reported that NQO1 knockout mice are more susceptible to development of DMBA-induced skin cancer (18). In addition, humans carrying the genetically polymorphic C609T NQO1 gene that encodes a biologically inactive enzyme show increased benzene toxicity (46) and are at an increased risk for development of different solid tumors and leukemias (19–22). Our results show that wild-type NQO1 stabilized wild-type p53 whereas the C609T polymorphic inactive NQO1 did not. It can be suggested that, after exposure to certain carcinogens that are substrates for detoxification by NQO1, cells that carry the defective gene would suffer a greater degree of damage but would express lower levels of p53 than normal cells. Because p53 is an important tumor-suppressor protein that can induce growth arrest and apoptosis, damaged NQO1-defective cells would have a viability and growth advantage that could lead to tumor development. For example, we have shown here that wild-type NQO1 partially antagonizes p53 degradation by the papillomavirus protein E6. Our data predict that papillomavirus-infected individuals who carry the inactive NQO1 gene will be more prone to p53 degradation and have a higher risk of developing cervical carcinoma. Epidemiological studies could clarify this point, which can have an important prognostic value.
We have shown previously that the NQO1 inhibitor dicoumarol induces degradation of mutant p53 in tumor cells and have suggested that dicoumarol might be used in combination with cytotoxic agents in therapy against tumors that express high levels of mutant p53 (17). Hsp90 inhibitors also caused degradation of mutant p53 in tumor cells but through a distinct pathway. Because hsp90 is also an effective suppressor of apoptosis by virtue of its ability to block activation of caspase 9 (47), the combination of hsp90 inhibitors and dicoumarol with cytotoxic agents could be used to improve therapy against tumors that express high levels of mutant p53.
Acknowledgments
We thank S. Budilovsky for her assistance; Dr. T. Unger for the pSG424 human p53 plasmid; Dr. M. Oren for the PAb 1801 anti-p53 antibody, the monoclonal anti-Mdm-2 antibody, and the pCOC-mdm2 X2 and pRc/CMV-E6 plasmids; Dr. U. Nudel for the pCGN-HA LT plasmid; and Dr. B. Vogelstein for the p53 null HCT116 cells. This work was supported by the Israel Academy of Sciences and Humanities and by research grants from the Benoziyo Institute of Molecular Medicine, the Dolfi and Lola Ebner Center for Biomedical Research, and the Otolaryngology Research Foundation.
Abbreviations
- HA
hemagglutinin
- hsp90
heat shock protein 90
- NQO1
NAD(P)H: quinone oxidoreductase 1
- SV40 LT
simian virus 40 large T antigen
- ROS
reactive oxygen species
- CMV
cytomegalovirus
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine A J. Nature (London) 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 4.Lotem J, Sachs L. Blood. 1993;82:1092–1096. [PubMed] [Google Scholar]
- 5.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 6.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 7.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 9.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Bayle J H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 11.Barak Y, Juven T, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Midgley C A, Lane D P. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 13.Peng Y, Chen L, Li C, Lu W, Chen J. J Biol Chem. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- 14.Blagosklonny M V, Toretsky J, Neckers L. Oncogene. 1995;11:933–939. [PubMed] [Google Scholar]
- 15.Dasgupta G, Momand J. Exp Cell Res. 1997;237:29–37. doi: 10.1006/excr.1997.3766. [DOI] [PubMed] [Google Scholar]
- 16.Nagata Y, Anan T, Yoshida T, Mizukami T, Taya Y, Fujiwara T, Kato H, Saya H, Nakao M. Oncogene. 1999;18:6037–6049. doi: 10.1038/sj.onc.1202978. [DOI] [PubMed] [Google Scholar]
- 17.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Proc Natl Acad Sci USA. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long D J, II, Waikel R L, Wang X-J, Roop D J, Jaiswal A K. J Natl Cancer Inst. 2001;93:1166–1170. doi: 10.1093/jnci/93.15.1166. [DOI] [PubMed] [Google Scholar]
- 19.Wiemels J L, Pagnamenta A, Taylor G M, Eden O B, Alexander F E, Greaves M F United Kingdom Childhood Cancer Study Investigators. Cancer Res. 1999;59:4095–4099. [PubMed] [Google Scholar]
- 20.Lafuente M J L, Casterad X, Trias M, Ascaso C, Molina R, Ballesta A, Zheng S, Wiencke J K, Lafuente A. Carcinogenesis. 2000;21:1813–1819. doi: 10.1093/carcin/21.10.1813. [DOI] [PubMed] [Google Scholar]
- 21.Smith M T, Wang Y, Kane E, Rollinson S, Wiemels J L, Roman E, Roddam P, Cartwright R, Morgan G. Blood. 2001;97:1422–1426. doi: 10.1182/blood.v97.5.1422. [DOI] [PubMed] [Google Scholar]
- 22.Lewis S J, Cherry N M, Niven R M, Barber P V, Povey A C. Lung Cancer. 2001;34:177–183. doi: 10.1016/s0169-5002(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 23.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 24.Lotem J, Sachs L. Proc Natl Acad Sci USA. 1977;74:5554–5558. doi: 10.1073/pnas.74.12.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalovitz D, Halevy O, Oren M. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 26.Barak Y, Gottlieb E, Juven Gershon T, Oren M. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 27.Matlashewski G, Banks L, Pim D, Crawford L. Eur J Biochem. 1986;154:665–672. doi: 10.1111/j.1432-1033.1986.tb09449.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Marechal V, Levine A J. Mol Cell Biol. 1993;13:4197–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lotem J, Peled-Kamar M, Groner Y, Sachs L. Proc Natl Acad Sci USA. 1996;93:9166–9171. doi: 10.1073/pnas.93.17.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross D, Traver R D, Siegel D, Kuehl B L, Misra V, Rauth A M. Br J Cancer. 1996;74:995–996. doi: 10.1038/bjc.1996.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegel D, Anwar A, Winski S L, Kepa J K, Zolman K L, Ross D. Mol Pharmacol. 2001;59:263–268. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- 32.Whitesell L, Sutphin P D, Pulcini E J, Martinez J D, Cook P H. Mol Cell Biol. 1998;18:1517–1524. doi: 10.1128/mcb.18.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 34.Lotem J, Sachs L. Apoptosis. 1999;4:187–196. doi: 10.1023/a:1009614723237. [DOI] [PubMed] [Google Scholar]
- 35.Oren M. J Biol Chem. 1999;274:36031–36034. doi: 10.1074/jbc.274.51.36031. [DOI] [PubMed] [Google Scholar]
- 36.de Oca Luna R M, Wagner D S, Lozano G. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 37.Jones S N, Roe A E, Donehower L A, Bradley A. Nature (London) 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 38.King F W, Wawrzynow A, Höhfeld J, Zylicz M. EMBO J. 2001;20:6297–6305. doi: 10.1093/emboj/20.22.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang N-S, Pratt N, Heath J, Schultz L, Sleve D, Carey G B, Zevotek N. J Biol Chem. 2001;276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 40.Dinkova-Kostova A T, Talalay P. Free Radical Biol Med. 2000;29:231–240. doi: 10.1016/s0891-5849(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 41.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. Nature (London) 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 42.Tan M, Li S, Swaroop M, Guan K, Oberley L W, Sun Y. J Biol Chem. 1999;274:12061–12066. doi: 10.1074/jbc.274.17.12061. [DOI] [PubMed] [Google Scholar]
- 43.Hwang P M, Bunz F, Yu J, Rago C, Chan T A, Murphy M P, Kelso G F, Smith R A J, Kinzler K W, Vogelstein B. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drane P, Bravard A, Bouvard V, May E. Oncogene. 2001;20:430–439. doi: 10.1038/sj.onc.1204101. [DOI] [PubMed] [Google Scholar]
- 45.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman W H, Tom E, Mack D H, Levine A J. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 46.Moran J L, Siegel D, Ross D. Proc Natl Acad Sci USA. 1999;96:8150–8155. doi: 10.1073/pnas.96.14.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandey P, Saleh A, Nakazawa A, Kumar S, Srinivasula S M, Kumar V, Weichselbaum R, Nalin C, Alnemri E S, Kufe D, et al. EMBO J. 2000;19:4310–4322. doi: 10.1093/emboj/19.16.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]