Abstract
We are developing methods to image molecular and cellular events in living subjects. In this study, we validate imaging of protein—protein interactions in living mice by using bioluminescent optical imaging. We use the well studied yeast two-hybrid system adapted for mammalian cells and modify it to be inducible. We employ the NF-κB promoter to drive expression of two fusion proteins (VP16-MyoD and GAL4-ID). We modulate the NF-κB promoter through tumor necrosis factor α. Firefly luciferase reporter gene expression is driven by the interaction of MyoD and ID through a transcriptional activation strategy. We demonstrate the ability to detect this induced protein–protein interaction in cell culture and image this induced interaction in living mice by using transiently transfected cells. The current approach will be a valuable and potentially generalizable tool to noninvasively and quantitatively image protein–protein interactions in living subjects. The approaches validated should have important implications for the study of protein–protein interactions in cells maintained in their natural in vivo environment as well as for the in vivo evaluation of new pharmaceuticals targeted to modulate protein–protein interactions.
The interaction of specific proteins in mammalian cells are essential for many biological processes. Protein–protein interactions modulate gene expression, signal transduction, intracellular transport pathways, as well as therapeutic and sometimes toxic effects of pharmaceuticals. Several approaches have been developed to study protein–protein interactions in living cells, including split-reporter approaches, where a reporter protein is reconstituted when two proteins interact (1), fluorescence resonance energy transfer in which energy transfer occurs only if the fluorescent labels of interacting proteins are in close proximity (2), as well as yeast two-hybrid approaches in which a reporter gene is primarily expressed only if two proteins interact (3, 4). The yeast two-hybrid system uses a reporter gene as a method for indirectly monitoring protein–protein interaction. Each of these approaches has distinct advantages (5), and, in the current work, we have adapted the yeast two-hybrid approach, usually used to screen for potential protein–protein interactions, for imaging known protein–protein interactions in living mice.
We and others have been developing reporter gene approaches suitable for use in living animals by using both positron-emission tomography (PET) (6–9, ‖) and optical bioluminescent approaches (10, 11). These approaches use reporter genes that encode for reporter proteins that can be detected in a living animal through injection of specific substrates (reviewed in ref. 12). Reporter genes with optical signatures (e.g., fluorescent and bioluminescent) are a low-cost alternative for real-time analysis of gene expression in small animal models. In fluorescent approaches (e.g., green fluorescent protein), an external source of light is required for excitation of the protein. In contrast, bioluminescent reporter proteins can produce light by using appropriate substrates. Recently, several technical advances in developing highly sensitive detection devices have led to the biological use of cooled charge-coupled device (CCD) cameras capable of imaging very low levels of visible light emitted from internal body organs of rodents (10, 13). A mouse can be placed in a light-tight box, and projection images of CCD-imaged bioluminescence can be superimposed on photographic images of the mouse to quantitatively and repetitively image the bioluminescent signal from a given location. We previously have validated the reproducibility and quantitative capability of this approach (10).
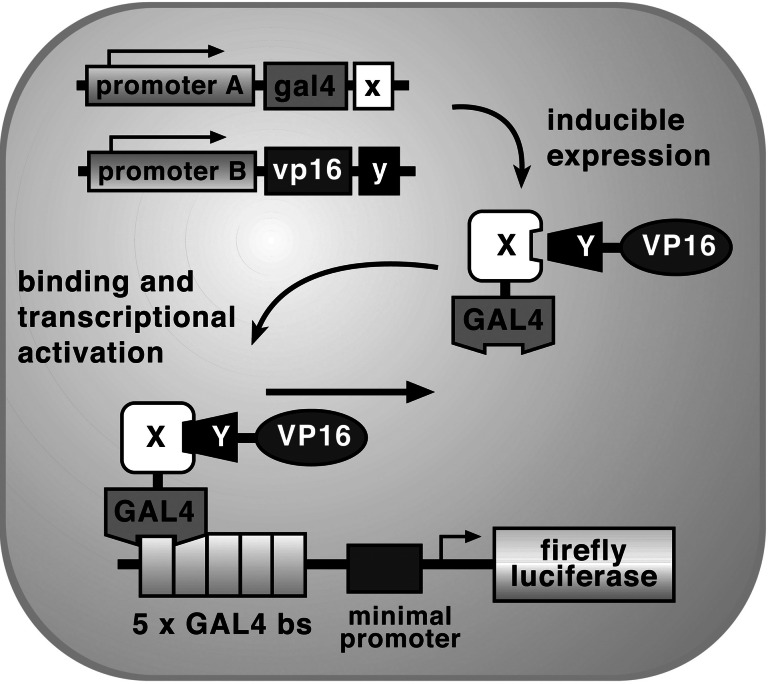
We recently validated the use of a two-step transcriptional amplification (TSTA) approach for enhancing reporter gene expression from relatively weak promoters in living mice (14). This system uses a GAL4-VP16 transactivation strategy to amplify expression of either the bioluminescent firefly luciferase (fl) or herpes simplex virus type 1 thymidine kinase (HSV1-tk) PET reporter genes. Significant reporter gene expression is generated only when the GAL4-VP16 fusion binds to GAL4-binding sites, leading to VP16-mediated transactivation of the reporter template. We also recently have refined this system further for even greater amplification through creating a single vector construct with potential feedback elements (15). We reasoned that the TSTA system could be modified so that indirect imaging of protein–protein interactions could be performed. In the current approach, the GAL4 and VP16 proteins are translated separately and are brought together through specific interactions of two proteins of interest X and Y (Fig. 1). We are able to modulate the production of the fusion proteins GAL4-X and VP16-Y, and the interactions of X and Y lead to the formation of the protein GAL4-X-Y-VP16, which is needed for transactivation of the reporter template. The reporter template contains five GAL4 DNA-binding sites and utilizes the fl reporter gene. (Note that fl refers to the gene and FL, to the enzyme.) Transcription of fl leads to FL, which is quantitatively imaged by injecting d-luciferin into the subject. d-Luciferin serves as a substrate for FL and leads to CCD-detectable bioluminescence (16). We term this approach an inducible yeast two-hybrid (IY2H) system.
Figure 1.
Schematic diagram of the IY2H system for imaging the interaction of proteins X and Y. The first step involves the vectors pA-gal4-x and pB-vp16-y, which are used to drive transcription of gal4-x and vp16-y through use of promoters A and B. In the second step, the two fusion proteins GAL4-X and VP16-Y interact because of the specificity of protein X for protein Y. Subsequently, GAL4-X-Y-VP16 binds to GAL4-binding sites [five GAL4-binding sites (bs) are available] on a reporter template. This leads to VP16-mediated transactivation of firefly luciferase reporter gene expression under the control of GAL4 response elements in a minimal promoter. Transcription of the firefly luciferase reporter gene leads to firefly luciferase protein, which, in turn, leads to a detectable visible light signal in the presence of the appropriate substrate (d-luciferin), ATP, Mg2+, and oxygen. In the current paper, we validate an inducible IY2H system while using the NF-κB promoter for either pA or pB and TNF-α-mediated induction. The use of the IY2H system allows indirect monitoring of protein–protein interactions through VP16-mediated transactivation of a reporter gene.
To validate the IY2H system, we used the two proteins ID and MyoD, which are known to strongly interact in vivo (17–19). MyoD normally is expressed in skeletal muscle and is a myogenic regulatory protein. The ID protein acts as a negative regulator of myogenic differentiation. MyoD and ID are members of the helix–loop–helix family of nuclear proteins. (Note that id-gal4 and myoD-vp16 refer to the fusion genes whereas ID-GAL4 and MyoD-VP16 refer to the fusion proteins.) To modulate the expression of these two proteins, we used the NF-κB promoter to drive expression of the id-gal4 and/or myoD-vp16 fusion genes while using tumor necrosis factor α (TNF-α) to induce the NF-κB promoter. TNF-α is a pleiotropic cytokine secreted by lipopolysaccharide-stimulated macrophages that induces a variety of cell-specific events and causes tumor necrosis in vivo when injected in tumor-bearing mice (20). The type 1 TNF-α receptor is a 55-kDa protein that is associated with a variety of functions when activated, including apoptosis, NF-κB activation, and Jun N-terminal kinase activation. The induction of both NF-κB activity and apoptosis by type 1 TNF-α receptor is mediated through its intracellular “death domain” region. In the TNF-α-mediated activation of NF-κB, a pathway is stimulated in which the last step is the phosphorylation-dependent degradation of IκB, the negative regulator of NF-κB, by proteosomes (21). We chose this promoter because it previously has been shown to be modulated in cell culture and in living animals with TNF-α.
We transiently transfected 293T cells with various combinations of plasmids first to verify fl expression in cell culture under various inducible and constitutive conditions along with the appropriate controls. We then performed cooled CCD imaging experiments in living mice implanted in the peritoneum with transiently transfected 293T cells to validate the ability to image protein–protein interaction in living mice by using TNF-α to modulate the system.
Materials and Methods
Chemicals.
TNF-α was purchased from Sigma, and Superfect transfection reagent was purchased from Qiagen. Luciferase assay kit was purchased from Promega, and d-luciferin for use with in vivo fl imaging was purchased from Xenogen (Alameda, CA). The polyclonal antibody against the GAL4 protein was a kind gift from M. Carey (University of California, Los Angeles).
Cell Culture.
Human embryonic kidney cancer cells, 293T (ATCC, Manassas, VA), were grown in MEM supplemented with 10% FBS and 1% penicillin/streptomycin solution. The C6 rat glioma cells were kindly provided by Margaret Black (Washington State University, Pullman) and were grown in deficient DMEM, supplemented with 5% FBS and 1% penicillin/streptomycin/l-glutamine. The N2a cells were obtained from V. P. Mauro (Scripps Research Institute, La Jolla, CA) and were grown in DMEM (high glucose) supplemented with 10% FBS and 1% penicillin/streptomycin.
Plasmid Construction.
All plasmids used are shown in Table 1. The PC, PD, and PR vectors were obtained from the CheckMate Mammalian Two-Hybrid system kit purchased from Promega. The PC vector contains the yeast GAL4 DNA-binding domain fused with the cDNA of ID protein, and the PD vector contains the HSV VP16 activation domain fused with a segment of murine MyoD cDNA. The PR vector contains five GAL4-binding sites upstream of a minimal TATA box, which, in turn, is upstream of fl. The PA and PB vectors were constructed to replace the constitutive cytomegalovirus (CMV) promoter with TNF-α-inducible NF-κB response elements (described below).
Table 1.
Plasmids used in study
| Nomenclature | Plasmid |
|---|---|
| PA | pNF-κB-gal4-id |
| PB | pNF-κB-vp16-myoD |
| PC | pCMV-gal4-id |
| PD | pCMV-vp16-myoD |
| PE | pCMV-fl |
| PF | pCMV-gal4-p53 |
| PR | pCMV-(gal4bs)5-fl |
The 194-bp segment consisting of a short, 39-bp NF-κB response element (kB4) and a 148-bp-long TATA-like promoter (PTAL) was excised from pNF-κB-Luc (CLONTECH) by KpnI and HindIII digestion and cloned in pBAD-MycHisA (Invitrogen) to generate proper restriction sites suitable for cloning in PC/PD vector. PC vector was digested completely with BglII and then partially with HindIII to excise the 750 bp of CMV promoter. To construct the PA plasmid, the pBAD-NF-κB-MycHisA was digested with BglII and HindIII to release the kB4-PTAL fragment, which then cloned into partially digested PC vector.
To construct the PB plasmid, we digested the pCMV-vp16 (available with the kit) vector completely with BglII and partially with HindIII to remove the CMV promoter. The same kB4-PTAL fragment released by BglII and HindIII digestion of pBAD-NF-κB-MycHisA vector then was inserted into the above-mentioned pCMV-vp16/BglII-HindIII fragment. The MyoD fragment from PD vector was released with BamHI and KpnI and finally cloned into the BamHI and KpnI sites of pNF-κB-vp16 plasmid to obtain the PB plasmid. The PF vector expresses the GAL4-binding domain and amino acids 72–390 of murine p53 as a hybrid protein and was obtained from the Mammalian Two-Hybrid Assay kit from Stratagene.
Cell Transfections and Luciferase Assay.
On day 1, 293T cells were plated in 12-well plates in MEM containing 10% FBS. Transient transfections were performed 24 h later by using Superfect transfection reagent. Each transfection mix consisted of 1.5 μg of the effector and reporter plasmids or reporter plasmid alone per well. Three hours after transfection, TNF-α was added to the medium at a concentration of 0.05 μg/ml and the cells were incubated for 24 h. The cells were harvested and assayed for FL activity by using the Dual-Reporter Luciferase Assay System (Promega) and a luminometer (Lumat 9507; Berthold, Nashua, NH) with an integration time of 10 sec. For TNF-α dose-response experiments, 293T cells were transiently transfected in the presence of different concentrations of TNF-α (0.005, 0.01, 0.05, and 0.1 μg/ml). After 24 h, the cells were assayed for FL activity. Identical studies were repeated by using a fixed concentration of 0.05 μg/ml TNF-α and varying times of exposure to TNF-α (0, 6, 8, and 24 h). The same experiments were repeated with two other cell lines (C6 and N2a) cultured in different media.
In Vivo Optical Imaging of fl Expression by Using a Cooled CCD Camera.
293T cells were transiently transfected with either plasmid PR alone or plasmids PA+PB+PR. The cells were harvested 3 h after transfection and resuspended in PBS. An aliquot of 1 × 106 cells was injected i.p. in anesthetized (ketamine/xylazine, 4:1) nude mice. Fifteen minutes after injection of the cells, 100 μl of d-luciferin (30 mg/ml) was injected 5 min before imaging. Throughout the study, d-luciferin and TNF-α were injected i.p. After the first scan, the mice were injected i.p. with 0.5 μg of TNF-α and imaged again after 8, 22, 30, and 48 h. Each time after scanning, the mice were reinjected with another dose of TNF-α. A total of six mice were used for the PA+PB+PR group that received TNF-α, and four mice were used for the PA+PB+PR group that did not receive TNF-α. Four control mice were injected i.p. with 293T cells transiently transfected with PR alone or PA+PF+PR and imaged repetitively over 24 h with (n = 2) and without (n = 2) repetitive injection of TNF-α every 8 h.
All mice were imaged by using a cooled CCD camera (Xenogen IVIS; Xenogen). The animals were placed supine in a light-tight chamber, and a gray-scale reference image was obtained under low-level illumination. Photons emitted from cells implanted in the mice were collected and integrated for a period of 2 min. Images were obtained by using living image software (Xenogen) and igor image analysis software (WaveMetrics, Lake Oswego, OR). For quantitation of measured light, regions of interest were drawn over the peritoneal region and maximum photons/sec per cm2 per steradian were obtained as validated previously (10).
Statistical Testing.
All cell culture and mouse group comparisons were performed with a Student's t test by using Microsoft excel 98. Values of P ≤ 0.05 were considered statistically significant.
Results
The IY2H System Mediates the Interaction of ID and MyoD Proteins as Measured by fl Reporter Gene Expression.
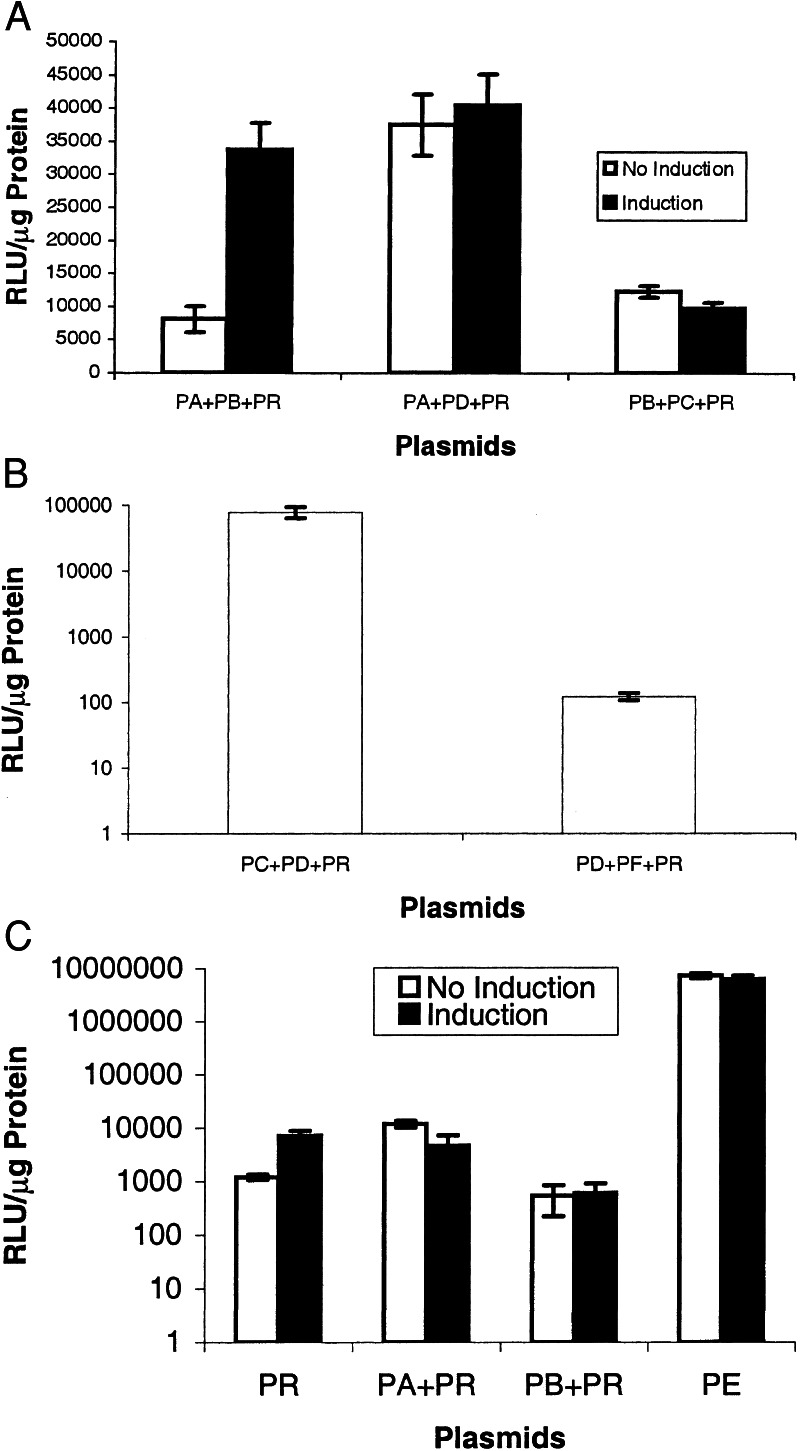
In transient transfection of plasmids PA+PB+PR into 293T cells, fl expression was relatively low in the absence of TNF-α but in its presence was significantly higher (P < 0.05) (Fig. 2A). The level of induction was ≈4-fold. When one of the two proteins (ID or MyoD) is constitutively expressed and the other is driven by NF-κB, there is no significant induction observed (Fig. 2A). Of note are the lower levels of fl expression when PB+PC vs. PA+PD are used. In transient transfection of plasmids PC+PD+PR into 293T cells, fl expression is relatively high and likely a result of the constitutive expression of GAL4-ID and VP16-MyoD proteins (Fig. 2B). In transient transfection of plasmids PD+PF+PR, there is relatively low fl expression because of the lack of significant interaction between MyoD and p53 proteins (Fig. 2B). The appropriate negative control studies also were performed by using transient transfection studies in 293T cells and plasmids PR alone, PA+PR, and PB+PR with and without TNF-α induction (Fig. 2C). The negative controls do not show any significant induction in fl expression pre- and post-TNF-α and show significantly lower (P < 0.05) fl expression than the postinduction values seen in Fig. 2A with PA+PB+PR. The positive control (PE alone) has a relatively high signal and does not show inducibility in the presence of TNF-α (Fig. 2C).
Figure 2.
(A) IY2H system-mediated fl expression. 293T cells were transiently transfected with (i) PA+PB+PR, (ii) PA+PD+PR, and (iii) PB+PC+PR. After 24 h in the absence and presence of TNF-α, the cells were harvested and assayed for FL activity. RLU, relative light units. Error bars represent SEM for triplicate measurements. (B) IY2H system with interacting and noninteracting protein partners. (Note logarithmic scale for y axis.) 293T cells were transiently transfected with plasmids (i) PC+PD+PR (interacting protein partners) and (ii) PD+PF+PR (noninteracting protein partners). After 24 h, the cells were harvested and assayed for FL activity. Error bars represent SEM for triplicate measurements. (C) Control cell culture studies. 293T cells were transiently transfected with (i) PR alone, (ii) PA+PR, (iii) PB+PR, and (iv) PE alone. After 24 h in the absence and presence of TNF-α, the cells were harvested and assayed for FL activity. (Note logarithmic scale for y axis.) Error bars represent SEM for triplicate measurements.
To show that the IY2H system is specifically inducible only in cells in which there are TNF-α receptors, plasmids PA+PB+PR were transiently transfected into 293T, C6, and N2a cells, and FL signal quantitated both with and without exposure to TNF-α. These studies show induction in 293T cells and lack of significant induction in the two other cell lines, demonstrating the requirement of the NF-κB signal transduction pathway for induction of the IY2H system. A Western blot analysis performed on protein extracted from 293T cells pre- and postinduction shows that there is presence of GAL4 protein and that it is relatively higher postinduction (data not shown). Furthermore, in 293T cells not transiently transfected, there is no band observed on the Western blot (data not shown).
Expression of fl for the IY2H system in the presence of TNF-α therefore is achievable in 293T cells. These results validate the concept that expression of both gal4-id and vp16-myoD leads to the production of GAL4-ID and VP16-MyoD proteins. The interaction of ID and MyoD then leads to transactivation of fl, allowing indirect confirmation regarding ID-MyoD interaction. Furthermore, the system requires the presence of both ID and MyoD and induction with TNF-α requires an intact NF-κB signal transduction pathway, as demonstrated with the control studies.
The IY2H System Can Be Modulated by TNF-α Concentration in Cell Culture.
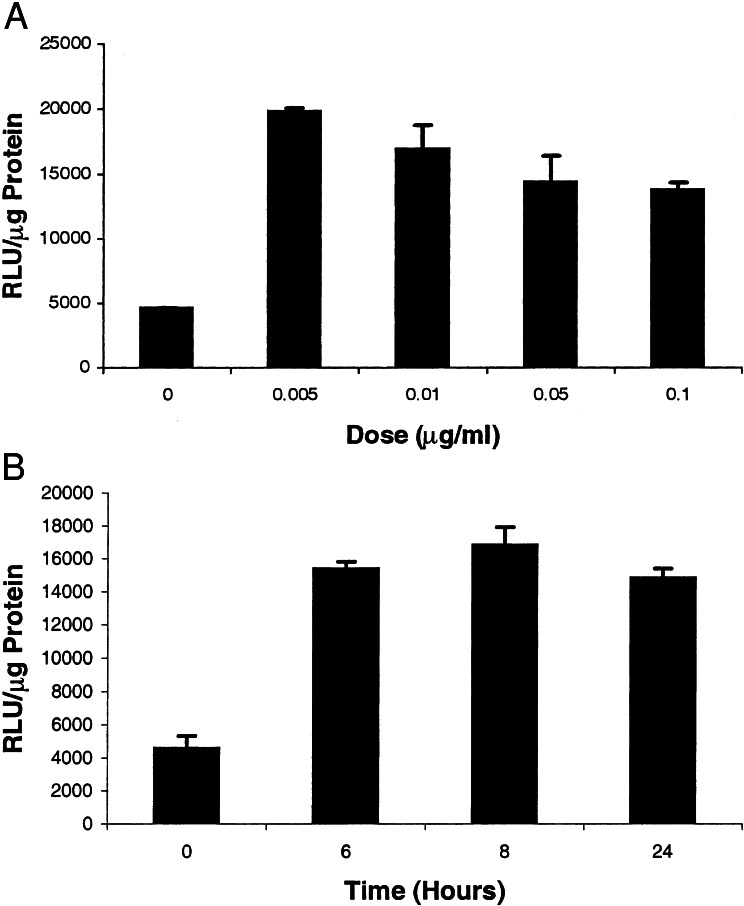
To test the ability to modulate the IY2H system, we transfected plasmids PA+PB+PR into 293T cells and exposed cells to increasing concentrations of TNF-α for a fixed time period of 24 h. Increasing levels of TNF-α led to increases in FL signal up to a concentration of 0.005 μg/ml, and then a progressive gradual decrease in FL signal was seen at higher concentrations (Fig. 3A). To study the time kinetics of TNF-α-mediated induction, the above studies were repeated with a fixed TNF-α concentration of 0.05 μg/ml and levels of FL signal were measured over a course of 24 h. These studies show that peak levels of FL are observed at ≈8 h after introduction of TNF-α (Fig. 3B) and that FL signal remains relatively fixed out to 24 h. These results demonstrate that the IY2H system can be modulated in a continuous fashion with increasing doses of TNF-α and that there is a time-dependent response that peaks at ≈8 h and then is relatively fixed out to 24 h.
Figure 3.
(A) Effect of TNF-α concentration on fl expression. 293T cells were transiently transfected with plasmids PA+PB+PR. The cells were harvested 24 h after transfection in the presence of different concentrations of TNF-α and assayed for FL activity. Error bars represent SEM for triplicate measurements. (B) Effects of TNF-α exposure time on fl expression. 293T cells were transiently transfected with plasmids PA+PB+PR. After a fixed time period of exposure to 0.05 μg/ml TNF-α, the cells were harvested and assayed for FL activity. Error bars represent SEM for triplicate measurements.
The IY2H System Can Be Used to Image Protein–Protein Interactions in Living Mice.
To test further the utility of the IY2H system in vivo, we injected transiently transfected (with plasmids PA+PB+PR, PB+PF+PR, or PR alone) 293T cells i.p. in nude mice. Mice injected i.p. with 293T cells transiently transfected with PA+PB+PR that are not induced with TNF-α show a relatively low but increasing fl expression over the course of 30 h (Fig. 4A). Mice injected i.p. with 293T cells transiently transfected with PA+PB+PR that are induced with TNF-α show increasing fl expression over 30 h (Fig. 4B). Mice injected i.p. with control 293T cells transfected with PR alone or PB+PF+PR (noninteracting proteins) show relatively low fl expression (<9 × 103 photons/sec per cm2 per steradian) from the peritoneum pre- and postinduction with TNF-α (data not shown). These results are indicative of the specificity of TNF-α in activating the NF-κB promoter, leading to production of ID and MyoD proteins and subsequent transactivation of the reporter template in vivo.
Figure 4.
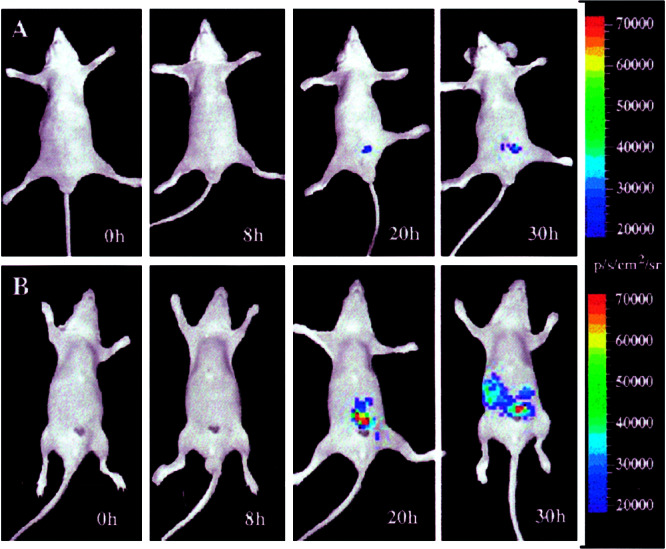
In vivo optical CCD imaging of mice carrying transiently transfected 293T cells for induction of the yeast two-hybrid system. All images shown are the visible light image superimposed on the optical CCD bioluminescent image with a scale in photons/sec per cm2 per steradian (sr). Mice are imaged in a supine position 5 min after injection of d-luciferin. (A) A nude mouse was imaged repetitively after being implanted with 293T cells transiently transfected with plasmids PA+PB+PR, and the mouse did not receive any TNF-α. There is some minimal gain in signal from the peritoneum over 30 h. Some other mice did show higher signals at later times. (B) A different nude mouse was imaged repetitively after being implanted with 293T cells transiently transfected with plasmids PA+PB+PR with TNF-α administration immediately after obtaining each image. There is a marked gain in signal from the peritoneum over 30 h. Note that the cells disperse from time 20 to 30 h, so the signal appears to come from a larger region. This particular mouse did not show as high a level of induction as other mice and showed induction relatively later at 20 h as opposed to other mice, which first showed induction at 8 h.
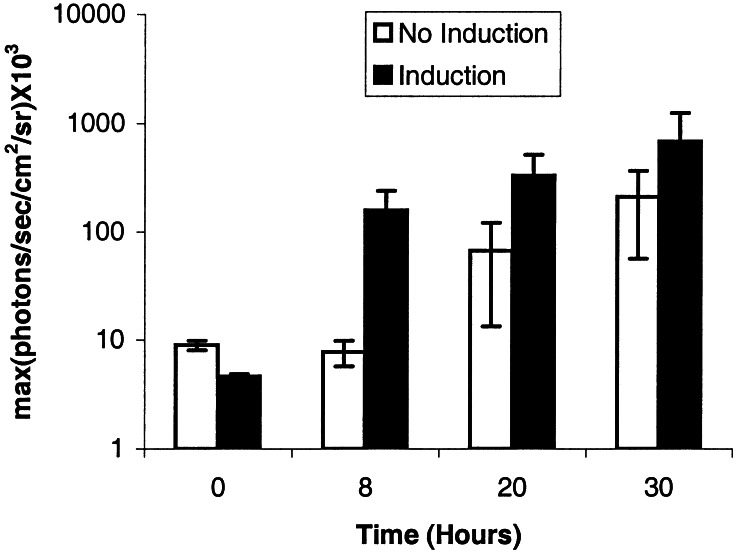
Transcriptional activation with (n = 6) and without (n = 4) TNF-α administration across 10 mice each injected i.p. with 1 × 106 293T cells transiently transfected with plasmids PA+PB+PR over the course of 30 h is illustrated in Fig. 5. IY2H system-mediated fl expression shows a significant gain (P < 0.05) at 8 h as compared with the uninduced group. At 20 and 30 h, there is greater induction in the induced group relative to the uninduced group but it is not statistically significant. There is significantly greater (P < 0.05) fl expression at 8, 20, and 30 h in the induced group as compared with preinduction levels at 0 h. There is a greater variability in vivo as compared with cell culture results. These results demonstrate the ability to image the interaction of two proteins in living subjects.
Figure 5.
Comparison of mice-imaging data with and without TNF-α induction. Mean [maximum (photons/sec per cm2 per sr)] in six induced mice and four uninduced mice as a function of time. (Note logarithmic scale for y axis.) All mice had i.p. injection of 1 × 106 293T cells transiently transfected with plasmids PA+PB+PR. The induced group shows a significantly greater signal (P < 0.05) at 8, 20, and 30 h as compared with preinduction levels at time 0. The induced group shows a significantly greater (P < 0.05) signal at 8 h as compared with the uninduced group. At 0 h, there is no significant difference between the induced and uninduced groups. Error bars represent SEM.
Discussion
We first show in cell culture that the IY2H systems show significant transactivation of fl expression only when there is coexpression of the genes coding for the two proteins ID and MyoD. Control studies using two noninteracting proteins (p53 and MyoD) show background levels of fl expression as do controls using the reporter template alone or one of the two proteins (ID and MyoD) without its corresponding interacting protein partner. TNF-α-mediated induction of the IY2H system shows fl expression that is time- and dose-dependent. C6 and N2a cells that lack TNF-α type 1 receptors also served as controls and do not show significant induction of fl expression in the presence of TNF-α.
We further tested the utility of the IY2H system in vivo to noninvasively and quantitatively image protein–protein interactions. To validate this system in vivo, several issues needed consideration. These included the development of (i) cell lines stably expressing the two effector and reporter constructs and (ii) construction of adenoviral or retroviral vectors containing all of the components of the system. Both approaches require considerable time before they can be tested in vivo. To develop an approach that can achieve quicker throughput and expedite the process of in vivo evaluation, we injected i.p. transiently transfected 293T cells in nude mice. The mice were imaged by using a sensitive, cooled CCD camera. The IY2H system was studied with and without TNF-α induction to enhance transcription from the NF-κB promoter. All IY2H mice displayed very low levels of fl expression immediately after cells were implanted. Eight hours after TNF-α administration, the mice representing the IY2H system showed a significantly greater level of fl expression when compared with mice that did not receive TNF-α. We observed relatively high levels of induction with an ≈20-fold (8 h), 5-fold (20 h), and 3-fold (30 h) gain for the IY2H system over the mice not receiving TNF-α. The level of IY2H-based fl expression in vivo is dependent on the pharmacokinetics of TNF-α availability to cells and likely is dependent on TNF-α dosage, frequency, and route of administration. In cell culture studies and in vivo, the peak induction is at 8 h. In cell culture at 24 h, there was an ≈4-fold gain for the induced vs. uninduced system (Fig. 2A), which is comparable to an ≈5-fold gain seen in vivo at 20 h (Fig. 5). Future studies will need to address the exact correlation between levels of TNF-α in blood/peritoneum and the levels of induction in vivo.
It would be desirable that the in vivo sensitivity of the IY2H system be high so that minimal levels of changes in protein–protein interaction can be detected in a living animal. We achieved an in vivo level of FL signal at 8 h for the induced system that was ≈20-fold greater than the identical, noninduced system and ≈60-fold greater than the system in which noninteracting protein partners (MyoD and p53) were transiently transfected. It would be desirable that even greater signal differences be achievable in vivo. The level of induction for the noninduced and induced system will depend, in part, on the leakiness of the promoter and the degree to which it can be induced by TNF-α or other factors. Future studies to further enhance the sensitivity potentially can rely on feedback-amplification strategies (15), more sensitive reporter genes, as well as the use of stable cell lines in which clones can be preselected with low levels of uninduced expression and high levels of induced expression. One disadvantage of using stable clones is the longer time it takes to isolate such clones, but, for cases in which prolonged studies of protein–protein interaction are needed, stable clones or transgenic models will be needed. Although we placed the transiently transfected 293T cells relatively close to the surface of the animal in the peritoneum, the cells actually can be placed anywhere within the mouse and enough light can be transmitted through tissues to be detected externally (11). The exact location of cells may be important for studying specific protein–protein interactions while cells are in their normal environment.
In the current work, we used three distinct vectors to test the IY2H system. In future applications, a single vector potentially can be used in which both the inducible systems and the reporter template are combined. With the use of appropriately placed multicloning sites, it should be possible to readily change the promoters, coding sequences for proteins of interest, as well as the reporter gene itself. This then would allow rapid testing of any two new proteins of interest. Although we used the firefly luciferase reporter gene, it is easily possible to use a PET reporter gene (22, 23), green fluorescent reporter gene (24), or other reporter genes (reviewed in ref. 12) with the appropriate instrumentation for imaging expression in living animals. It also should be possible to use fusion reporters (e.g., luciferase fused with a PET reporter gene) to perform imaging with multiple modalities on the same living animal. The relative merits for each of the technologies especially in terms of in vivo sensitivity will require further investigation.
Both components of our IY2H system were induced by the use of the NF-κB promoter. Even without induction with TNF-α, the NF-κB promoter shows transcriptional activity as evidenced by increasing fl expression over time. When expression of both protein-coding sequences is under control of the CMV promoter, then fl expression is the greatest. Although the current system was validated with the CMV and NF-κB promoters, in future applications, it may be desirable to use promoters that normally regulate expression of the coding region of the proteins of interest. It also may be important to be able to have the protein concentrations in a range that is near-equivalent to their normal ranges so that protein–protein interactions are not biased because of nonphysiological levels of protein concentrations.
Several limitations of the current approach merit some discussion. The current system cannot fully identify the time kinetics of protein–protein interaction. If two proteins interact for some time and then stop, it still may be the case that fl expression will persist even when the two proteins are no longer interacting. This is due to an inherent time lag in the IY2H system because it is only after sufficient levels of both proteins exist that one achieves transactivation of the reporter template. Future studies to look at the half-lives of the fusion proteins, reporter mRNA, and protein will help to better characterize the full kinetics of the current system. Alternate approaches that reconstitute a reporter protein directly through the interaction of two proteins of interest may allow more direct kinetic measurements of protein–protein interactions (5). It also should be noted that the current system is dependent on the two proteins of interest interacting in the vicinity of the reporter template. If the two proteins interact, but not in a location near the reporter template, then there may not be any transactivation.
Direct extensions of the current work should allow novel investigations of protein–protein interactions in living subjects. Studies of protein–protein interaction already have evolved significantly from studies in vitro to those in living cells. The uses of the current approaches would not be to screen for potential protein interactions, but to take an existing protein–protein interaction and then study it in detail in the normal cellular environment of a living subject. The advantage of this methodology is that it would allow extracellular factors that may modulate protein–protein interactions to be studied carefully. Furthermore, pharmaceuticals that can modulate protein–protein interaction now potentially can be tested directly in living subjects.
Acknowledgments
We thank David Stout and Xiaoman Lewis for technical assistance. This work is supported in part by National Institutes of Health Grants P50 CA86306 (S.S.G.), R0–1 CA82214 (S.S.G.), and SAIRP R24 CA92865 (S.S.G.), Department of Energy Contract DE-FC03–87ER60615 (S.S.G.), and the Association for the Cure of Cancer of the Prostate (S.S.G.).
Abbreviations
- fl
firefly luciferase reporter gene
- FL
firefly luciferase enzyme/protein
- CCD
charge-coupled device
- PET
positron-emission tomography
- IY2H
inducible yeast two-hybrid system
- CMV
cytomegalovirus
- TNF-α
tumor necrosis factor α
Footnotes
Tjuvajev, J. G., Avril, N., Safer, M., Joshi, R., Oku, T., Sasjima, T., Miyagawa, T., Beattie, B., Daghighian, F., Augenson, F., et al. (1997) J. Nucl. Med. 38, 239P (abstr.).
References
- 1.Rossi F, Charlton C A, Blau H M. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams S, Harootunian A, Buechler Y, Taylor S, Tsien R. Nature (London) 1991;49:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 3.Bai C, Elledge S J. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 4.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 5.Johnsson N, Varshavsky A. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambhir S S, Barrio J, Wu L, Iyer M, Namavari M, Satyamurthy N, Bauer E, Parrish C, MacLaren D, Borghei A, et al. J Nucl Med. 1998;39:2003–2011. [PubMed] [Google Scholar]
- 7.Gambhir S S, Barrio J R, Phelps M E, Iyer M, Namavari M, Satyamurthy N, Wu L, Green L A, Bauer E, MacLaren D C, et al. Proc Natl Acad Sci USA. 1999;96:2333–2338. doi: 10.1073/pnas.96.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tjuvajev J G, Avril N, Oku T, Sasajima T, Miyagawa T, Joshi R, Safer M, Beattie B, DiResta G, Daghighian F, et al. Cancer Res. 1998;58:4333–4341. [PubMed] [Google Scholar]
- 9.MacLaren D C, Gambhir S S, Satyamurthy N, Barrio J R, Sharfstein S, Toyokuni T, Wu L, Berk A J, Cherry S R, Phelps M E, et al. Gene Ther. 1999;6:785–791. doi: 10.1038/sj.gt.3300877. [DOI] [PubMed] [Google Scholar]
- 10.Wu J C, Sundaresan G, Iyer M, Gambhir S S. Mol Ther. 2001;4:297–306. doi: 10.1006/mthe.2001.0460. [DOI] [PubMed] [Google Scholar]
- 11.Bhaumik S, Gambhir S S. Proc Natl Acad Sci USA. 2002;99:377–382. doi: 10.1073/pnas.012611099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray P, Bauer E, Iyer M, Barrio J R, Satyamurthy N, Phelps M E, Herschman H, Gambhir S S. Semin Nucl Med. 2001;31:312–320. doi: 10.1053/snuc.2001.26209. [DOI] [PubMed] [Google Scholar]
- 13.Contag C H, Jenkins D, Contag P R, Negrin R S. Neoplasia. 2000;2:41–52. doi: 10.1038/sj.neo.7900079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer M, Wu L, Carey M, Wang Y, Smallwood A, Gambhir S S. Proc Natl Acad Sci USA. 2001;98:14595–14600. doi: 10.1073/pnas.251551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, J., Adams, J., Billick, E., Ilagan, R., Iyer, M., Smallwood, A., Gambhir, S. S., Carey, M. & Wu, L. (2002) Mol. Ther., in press. [DOI] [PubMed]
- 16.Contag P R, Olomu I N, Stevenson D K, Contag C H. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 17.Finkel T, Duc J, Fearon E R, Dang C V, Tomaselli G F. J Biol Chem. 1993;268:5–8. [PubMed] [Google Scholar]
- 18.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, et al. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 19.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 20.Boland M P, O'Neill L A. J Biol Chem. 1998;273:15494–15500. doi: 10.1074/jbc.273.25.15494. [DOI] [PubMed] [Google Scholar]
- 21.Saltzman A, Searfoss G, Marcireau C, Stone M, Ressner R, Munro R, Franks C, D'Alonzo J, Tocque B, Jaye M, et al. FEBS Lett. 1998;425:431–435. doi: 10.1016/s0014-5793(98)00287-7. [DOI] [PubMed] [Google Scholar]
- 22.Gambhir S S, Barrio J R, Herschman H R, Phelps M E. Nucl Med Biol. 1999;26:481–490. doi: 10.1016/s0969-8051(99)00021-9. [DOI] [PubMed] [Google Scholar]
- 23.Herschman H, MacLaren D C, Iyer M, Namavari M, Bobinski K, Green L A, Wu L, Berk A J, Toyokuni T, Barrio J R, et al. J Neurosci Res. 2000;59:699–705. doi: 10.1002/(SICI)1097-4547(20000315)59:6<699::AID-JNR1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang M, Hasegawa S, Jiang P, Wang X O, Tan Y Y, Chishima T, Shimada H, Moossa A R, Hoffman R M. Cancer Res. 1998;58:4217–4221. [PubMed] [Google Scholar]