Abstract
We constructed a three-dimensional model of the amino-terminal extracellular domain of three major types of nicotinic acetylcholine receptor, (α7)5, (α4)2(β2)3, and (α1)2β1γδ, on the basis of the recent x-ray structure determination of the molluscan acetylcholine-binding protein. Comparative analysis of the three models reveals that the agonist-binding pocket is much more conserved than the overall structure. Differences exist, however, in the side chains of several residues. In particular, a phenylalanine residue, present in β2 but not in α7, is proposed to contribute to the high affinity for agonists in receptors containing the β2 subunit. The semiautomatic docking of agonists in the ligand-binding pocket of (α7)5 led to positions consistent with labeling and mutagenesis experiments. Accordingly, the quaternary ammonium head group of nicotine makes a π-cation interaction with W148 (α7 numbering), whereas the pyridine ring is close to both the cysteine pair 189–190 and the complementary component of the binding site. The intrinsic affinities inferred from docking give a rank order epibatidine > nicotine > acetylcholine, in agreement with experimental values. Finally, our models offer a structural basis for potentiation by external Ca2+.
The nicotinic acetylcholine receptors (nAChRs) are well characterized transmembrane allosteric proteins involved in fast ionic responses to acetylcholine (1, 2). They are composed of five identical (homopentamers) or different (heteropentamers) polypeptide chains arranged symmetrically around an axis perpendicular to the membrane. The agonist-binding sites are located at the interface between two subunits. Sequence analyses further reveal that the numerous nAChR subunits are homologous proteins that belong, together with the GABAA,C, glycine, and 5-HT3 receptors, to the “cys-loop” superfamily of ligand-gated ion channels (3–5). In vertebrates, the combinatorial assembly of the subunits (α1–10, β1–4, γ, δ, ɛ) generates a wide diversity of receptors, with various electrical and binding properties. nAChRs may spontaneously exist under different conformational states, basal or resting (closed), active (open), or desensitized (closed) (6). The presence of nicotinic ligands, agonists, or competitive antagonists, and also of noncompetitive allosteric effectors, affects the equilibrium between the various states. Indeed, the ligands will stabilize the states for which they display the highest affinity. In particular, agonists exhibit a higher affinity for the active than for the basal state, therefore causing the opening of the channel, and generally exhibit an even higher affinity for the desensitized states, causing the progressive fading of ionic fluxes. In contrast, snake toxins block the receptor by stabilizing the basal state (refs. 7–9; see also Fruchart-Gaillard et al., 10). Their extremely high affinity and selectivity permitted the first isolation and purification of the receptor (11).
Because of their functional importance and their implication in numerous pathologies (12, 13), the nAChRs have been thoroughly investigated. Two decades of site-directed mutagenesis and affinity labeling provided an impressive amount of data about the structure of the receptors (14, 15). Numerous spectroscopic studies (in particular circular dichroism) were also conducted on the recombinant amino-terminal extracellular domain of various nAChR subunits (16–19). These studies showed that the extracellular domain of nAChRs contains a high proportion of residues belonging to β-strands. In addition to these experimental works, secondary structure predictions suggested an immunoglobulin-like folding, almost exclusively composed of β-strands, for the amino-terminal domain (15, 20).
However, the three-dimensional structure of the nAChRs at the atomic level remained unknown due to the difficulty in crystallizing integral membrane proteins and to their size, too large to easily use the NMR approach. Direct structural studies were thus limited to electron microscopy diffraction on the receptor of the Torpedo electric organ, down to 4.6 Å resolution (21) at best. The situation changed dramatically with the recent x-ray determination at 2.7 Å resolution of the Acetylcholine-Binding Protein (AChBP; Protein Data Bank ID code 1I19) (22). AChBP is a soluble homopentameric homologue of the amino-terminal extracellular domain of nAChR, synthesized in glial cells of the snail Lymnaea stagnalis and released in the synaptic cleft, where it acts as a cholinergic buffer and regulates interneuronal transmission (23).
AChBP exhibits a significant sequence similitude with the extracellular parts of the nAChR subunits (Fig. 1). In particular, its sequence identity to the subunit α7 is above 26% (Table 1). In addition, the AChBP pentamer exhibits pharmacological properties roughly similar to the α7 homopentamer, albeit with a higher affinity for most of the ligands. These facts strongly suggest that AChBP is homologous to the amino-terminal portion of the nicotinic receptors, and that these proteins are similarly folded (24, 25).
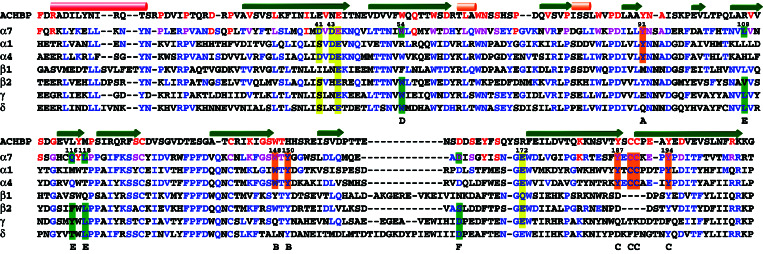
Figure 1.
Sequence alignment between AChBP and amino-terminal domains of chick α7, rat α4 and β2, and Torpedo α1, β1, γ, and δ nAChR subunits. The first line presents the secondary structure of AChBP, as determined with the program stride (26). The red rod represents an α-helix, whereas the orange rods represent 310 helices. The green arrows represent β-strands. The red residues are common to AChBP and α7, and the blue residues are common to α7 and another nAChR subunit (pink residues are common to AChBP, α7 and another nAChR subunit). Residues of the main component of the ACh-binding site are highlighted in orange, and those of the complementary component are highlighted in green, whereas the residues highlighted in yellow putatively belong to a Ca2+ allosteric site. Letters on the last line label the residues belonging to the agonist-binding site. The numbering is that of chick α7.
Table 1.
Percentage of sequence identities computed from the alignment shown in Fig. 1
| AChBP | α7gg | α4rn | β2rr | α1tc | γtc | δtc | |
|---|---|---|---|---|---|---|---|
| α7gg | 26.4 | ||||||
| α4rn | 21.7 | 45.6 | |||||
| β2rr | 19.7 | 40.6 | 54.7 | ||||
| α1tc | 20.8 | 38.3 | 52.7 | 49.3 | |||
| γtc | 20.9 | 34.8 | 39 | 47.5 | 39.3 | ||
| δtc | 22.7 | 35 | 39.4 | 44 | 38.3 | 53.9 | |
| β1tc | 22.6 | 35.1 | 41.7 | 43 | 40.5 | 49.5 | 47.0 |
gg, Gallus gallus; rn, Rattus norvegicus; rr, Rattus rattus; tc, Torpedo californica.
In this paper, we present a three-dimensional model of the extracellular domains of the three main types of nicotinic receptors, putatively in a conformation corresponding to a desensitized state. The homomeric chick nicotinic receptor (α7)5 was modeled on the basis of the crystal structure of AChBP. The heteromeric neuronal rat (α4)2(β2)3 and the heteromeric muscle Torpedo (α1)2β1γδ were then modeled on the basis of the (α7)5 model. We docked small agonists in the ligand-binding site and identified a putative structural determinant of the differential potentiation by external Ca2+.
Methods
Sequence Alignments.
The sequence alignment between AChBP and the amino-terminal domain of the chick α7 subunit was deduced from a sequence–structure alignment performed with the program deep-view (27). The resulting alignment exhibits small differences in five different regions with the alignment presented in ref. 22. Consequently, the identities climb from 22.3% for ref. 22 to 26.4%, at the cost of an extra gap. The multiple alignment between the amino-terminal domains of the chick α7, the rat α4 and β2, and the Torpedo α1, β1, γ, and δ was initially obtained with clustalx (28). This alignment was further manually refined to cope with the goal of comparative modeling.
Molecular Modeling.
The three-dimensional models were performed with the new version (version 6) of the program modeller (29), kindly provided by Andras Fiser (Rockefeller University). The automatic script “model” was used together with a routine to patch the two disulfide bonds of each subunit. The portions of the subunits that were modeled correspond to residues F2-T207 of the chick α7 subunit (Fig. 1). The buried surface and the intersubunit hydrogen bonds were determined with the Protein–Protein Interaction Server (http://www.biochem.ucl.ac.uk/bsm/PP/server/) (30). Cα rms deviation (rmsd) values, the average distance between homologous α carbons, were obtained in deep-view after superposing the whole structures or binding only one site of each molecule. Energy minimizations performed in deep-view with gromos96 parameters (http://igc.ethz.ch/gromos) did not significantly modify the initial models (all atom rmsd values inferior to 0.1 Å).
The docking of ACh, nicotine, and epibatidine was performed in two steps. First, the ligands were arbitrarily positioned in the agonist-binding pocket of the receptor by using deep-view. The model was previously protonated and the partial charges added (by a custom program given by Marc Delarue, Institut Pasteur), as well as the solvation parameters. Then the plausible dockings were found with the widely used program autodock3 (31). This program starts with a ligand molecule in an arbitrary conformation, orientation, and position and finds favorable dockings in a protein-binding site, using both simulating annealing and genetic algorithms. Interaction energies are calculated with a free-energy–based expression comprising several terms (dispersion/repulsion energy, directional hydrogen bonding, screened Coulomb potential electrostatics, a volume-based solvation term, and a weighted sum of torsional degrees of freedom to estimate the entropic cost of binding). The force field was calibrated with chemically diverse protein/ligand complexes of known structure.
Results
Model of Chick α7 Pentamer.
The use of several independent third-generation algorithms of secondary structure prediction suggested that the amino-terminal domain of nAChR subunits was almost exclusively composed of β-strands (20). The consensus prediction of a typical nAChR subunit amino-terminal domain exhibited 61.2% identity with the secondary structure of AChBP. The sequence identity between AChBP and α7, being 26%, implies a secondary structure identity around 80% between the two proteins (24), which in turn means that the true accuracy of our prediction could be a little higher (in fact, the identity of the prediction with the model α7 presented below is 63.6%). On the basis of the secondary structure prediction and other predicted properties, such as solvent accessibility, but also of experimental data, we proposed that this amino-terminal moiety was folded as a β sandwich, probably an immunoglobulin-like domain (15). The structure of the AChBP monomer effectively comprises an immunoglobulin fold (22).
We used the approach of comparative modeling based on a template of known structure. This approach is still the best available when a homolog of the target is known, even if the ab initio methods displayed a little improvement over the past years (32). It has already been used to successfully predict the structure of ligand-gated ion channels (33) and other neuronal proteins (34). However, it is important to keep in mind the limitations inherent in a modeling process built on a sequence identity inferior to 30% between the template and the target (35).
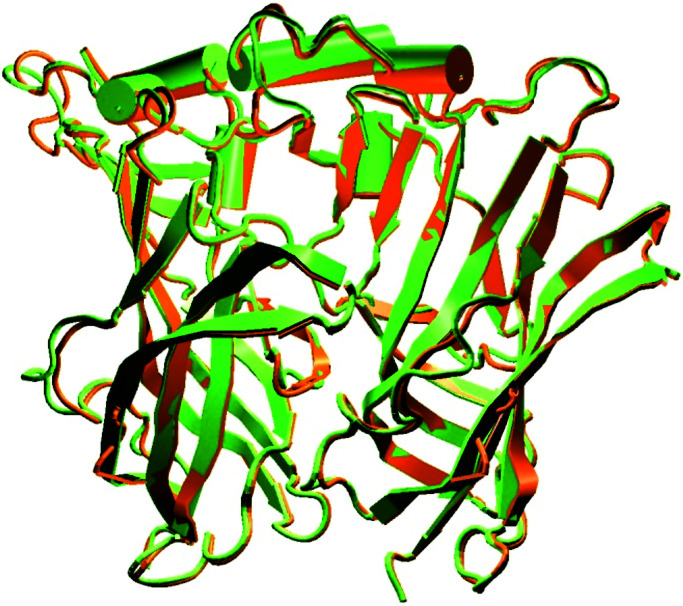
As expected from the low number of insertions/deletions in the alignment, and because of the algorithm used by modeller, the model of the amino-terminal portion of α7 does not exhibit major differences with the template (Fig. 2). The Cα rmsd is 0.71 Å. This value is low, although in the range expected for 25% identity, (0.7–2.3 Å) (24), suggesting that the replacement of three-fourths of the residues of AChBP with those of α7 did not introduce important constraints on the formation of secondary structure elements or on their interactions within the β sandwich. The model is shaped like a barrel, which, as described in Brejc et al. (22), looks like a toy windmill when viewed along the axis. Its diameter is 80 Å, whereas its height is 63 Å. The diameter of the central irregular pore ranges from 10 to 15 Å. Although the model is a homopentamer, the subunits are all slightly different, because of independent reconstruction by the modeling process. This is likely to represent the physiological situation, where the residues of each subunit explore the available conformers in an uncoupled manner. Moreover, modeling en bloc was judged preferable to a modeling of only one subunit followed by 5-fold symmetry. Indeed, the interfaces between subunits add constraints to the final model.
Figure 2.
Superposition of two adjacent subunits of AChBP (green) and two adjacent subunits of the model of α7 (orange). The figure was constructed with vmd (36) and rendered with raster3d (37).
The surface buried between two adjacent subunits is 1,250 Å2, with four hydrogen bonds crossing the interface. If we denote by (+) the surface of the subunit clockwise to the interface when the receptor is viewed from the extracellular space and by (−) the surface of the adjacent subunit, these bonds are (+)Y150–(−)R78, (+)W148–(−)L118, (+)D88–(−)N106, and (+)T149–(−)N106. The two former bonds are also present in AChBP on different residues located at the same place, which reinforces the plausibility of the model.
Several small pieces of α7 do not display any sequence identity with AChBP. The expected accuracy of the model in those regions is therefore lower than for the general structure. In particular, the 13-residue cys-loop, conserved across all the receptors of the superfamily and essential to their folding (38), is highly divergent in AChBP.
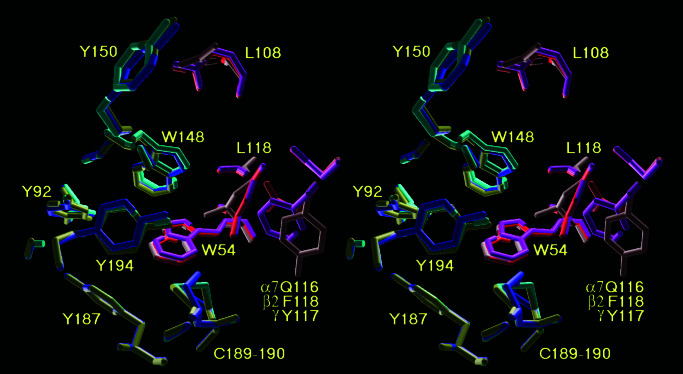
As for AChBP, nearly all the residues of the agonist-binding site identified by photoaffinity labeling and mutagenesis experiments are located in a small cavity of about 10- to 12-Å diameter (see Fig. 4). This cavity is formed by nine residues, mainly aromatic, located at the interface between two adjacent subunits. Seven residues form the “principal component” of the binding site: Y92 (loop A), W148, Y150 (loop B), and Y187, C189, C190, and Y194 (loop C). This principal component is located on the (+) surface of the interface. The box is completed by four residues located on the (−) surface of the interface: W54 (loop D), L108, Q116, and L118 (loop E), forming the “complementary component.” Note that three of these residues also form the hydrogen bonds linking adjacent subunits. As in AChBP, the residues W85 of the principal component (loop A) and D163 of the complementary component (loop F) do not participate in the agonist-binding site in the conformation modeled here. However, AChBP was crystallized in a conformation that corresponds to a still-undefined state of nAChR. From this state, clockwise rotations and/or tilting movements of all subunits could bring D163 closer to the agonist-binding site in alternative states.
Figure 4.
Stereoscopic representation of the superposed agonist-binding sites between two α7 subunits (blue and red), α4 and β2 (cyan and purple), and α1 and γ (yellow and grey).
Docking of the Small Agonists.
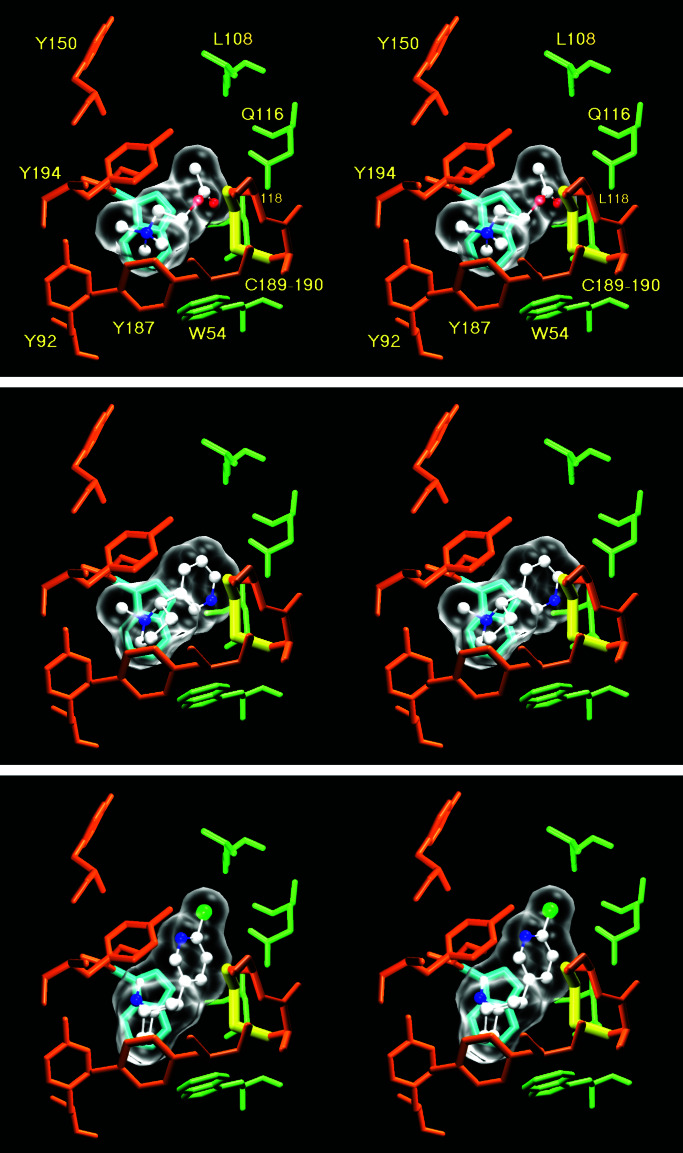
We used the program autodock3 to dock the small agonists acetylcholine, epibatidine, and nicotine on the model of α7. We manually positioned the ligands arbitrarily in the binding pocket and then ran up to 50 docking procedures per ligand. For each ligand, two or three clusters of positions were found. In each case, only one of the dockings agreed with the biochemical experiments (Fig. 3) performed on Torpedo nAChR. For instance, the methyl groups substituting the ammonium of ACh are close to Y92, as shown by Cohen et al. (39), whereas the ester bond of ACh was close to the cystine and to Y194, as shown by Grutter et al. (40). In addition, in the alternative positions, the three ligands were slightly shifted out of the binding pocket and contacted residues of the receptor that have never been identified by affinity labeling.
Figure 3.
Stereoscopic representation of the dockings of acetylcholine (Top), nicotine (Middle), and epibatidine (Bottom) onto the model of the amino-terminal part of α7. Oxygen atoms are red spheres, whereas nitrogens are blue, carbons are white, and the chlorine is green. The C189–190 disulfide bond is highlighted in yellow. W148, which makes a π-cation interaction with the ammonium, is highlighted in cyan. Note that all the sidechains are in the same position. Indeed, autodock does not allow sidechain movements during the docking process.
The selected position was common for the three ligands (whereas the alternative positions were different for each ligand). In particular, the ammonium of the three agonists was almost superposed in the chosen dockings, their distance being 0.78, 0.82, and 0.95 Å. This location is close to the position of an ammonium of the hepes molecule cocrystallized with AChBP, their distances being 0.82, 1.2, and 1.67 Å. In the selected dockings, the ammonium of the three ligands was able to establish a π-cation interaction with W148, as shown by Zhong et al. (41). Moreover, equivalent positions were retrieved when the docking procedure was performed by using binding pockets located between different subunits of the model of α7.
The Ki inferred from the estimated free energy of binding were in the range of tens of micromolar for acetylcholine, micromolar for nicotine, and hundredths of nanomolar for epibatidine. Those values not only reproduce the experimental ranking of binding efficiencies but also are in the same range, the measured Ki being 2.5⋅10−5 ± 0.5 M for acetylcholine, 0.55⋅10−6 ± 0.02 M for nicotine (42), and 1.9⋅10−7 ± 0.1 M for epibatidine (43). The binding free energy computed by autodock has been shown to reproduce correctly the experimental values (44). However, because of uncertainty on the comparative modeling process, absolute values have to be considered with caution at this stage.
α-Bungarotoxin and Allosteric States.
The high affinities mentioned above correspond to those measured at equilibrium on nAChRs in a desensitized state. This observation is consistent with the hypothesis that AChBP was crystallized in a compact conformation that corresponds to a desensitized state for nAChR (45). A model of interaction between α-bungarotoxin and AChBP has been presented by Harel et al. (46), on the basis of the crystallized—compact—form of AChBP. However, it has been shown by independent approaches that snake toxins block the receptor by stabilizing the basal state (7–9). In agreement with this scheme, Harel's model exhibits numerous clashes between the toxin and the subunit carrying the complementary component of the binding site. Accordingly, the toxin could recognize neither the conformation of AChBP that has been crystallized nor our own models of nAChRs, presented in the present article, but rather another conformation carrying a more opened agonist-binding site. The accompanying paper by Fruchart-Gaillard et al. (10) presents a model of α7 with an agonist-binding site modified to accommodate the α-cobratoxin. When we dock the small ligands on this model, we get significantly lower affinities, 100-fold for epibatidine and 10-fold for nicotine and acetylcholine. This important decrease of affinity supports the possibility that such a model of the binding site would be close to the conformation displayed in the basal rather than in the desensitized state.
Models of Rat (α4)2(β2)3 and Torpedo (α1)2β1γδ.
Because of the rather low sequence identity between AChBP and α4, β2, α1, β1, γ, or δ, the rat (α4)2(β2)3 and Torpedo (α1)2β1γδ were modeled by using the (α7)5 model as a template rather than AChBP directly. The sequence identities between the amino-terminal parts of α7 and its homologs range from 35 to 45%, which should result in a rmsd ranging from 0.6 to 1.9 Å (24). Indeed, the Cα rmsd between (α7)5 and (α4)2(β2)3 is 0.57 Å, whereas the Cα rmsd between (α7)5 and (α1)2β1γδ is 0.66 Å.
The Cα rmsd computed after superposition of the binding residues is lower than the rmsd computed after superposition of the global structures: 0.18 Å between AChBP and (α7)5, and 0.15 Å between (α7)5 and (α4)2(β2)3 and between (α7)5 and (α1)2β1γδ. This conservation of the binding residues is not solely due to the conservation of the residues, because three sets of nine randomly chosen conserved residues (index computer-generated) gave rmsds of 1.67, 0.63, and 0.78 Å. Moreover, not only is the position of the residues conserved within a subunit, but the distances between residues belonging to two adjacent subunits are also constant. The evolutionary constraints on the agonist-binding site could, therefore, be more important than the constraints on the overall structure.
The two binding sites of the Torpedo receptor are different, one being formed by an α and a γ subunit, and the other by an α and a δ subunit. These two binding sites have different affinities for several ligands. In particular, some competitive antagonists (47, 48) bind more strongly to the αγ-binding site. One of the residues involved in this differential affinity is γY117/δT119, located on the loop E (49). Fig. 4 shows that the tyrosine aryl faces the binding pocket and could reinforce the interaction with the ligand and therefore decrease the free energy of the complex. The same difference has been shown for the affinity towards epibatine of the mouse αγ- and αδ-binding sites (50).
Y117 of γ is a homolog of a phenylalanine in β2 and a glutamine in β4. Accordingly, regardless of the α subunit, the affinity for nicotine of receptors containing β4 is lower than one of the receptors containing β2. The loop E has already been suggested to be involved in this difference (51). In particular, the graft of β2 residues 104–120 confers β2-like affinities to β4 (note, however, that the reverse experiment was much less efficient; see below the work of Parker et al., ref. 52). We propose that the phenylalanine would be involved in the high affinity for nicotine of the desensitized state of β2-containing receptors. Recently, Parker et al. (52) showed that some residues of β4, flanking the binding tryptophan of loop D, transferred a lower affinity for nicotine to β2. The strongest, though moderate, effect (7- to 8-fold) was obtained with the mutation T59K. However, in our (α4)2(β2)3 model, the threonine makes direct contact not with the docked ligands but rather with the phenylalanine of loop E, suggesting that the mutation would allosterically affect the agonist site. In agreement with this observation, the mutant T59D, carrying an opposite charge, did produce a similar loss of affinity.
Effect of [Ca2+]o on the Allosteric Constant.
The response of nAChRs is modulated by a variety of allosteric effectors (53). In particular, extracellular calcium potentiates several nAChRs both in vivo (54, 55) and in transfected cells (56, 57).
Galzi et al. (57) identified by site-directed mutagenesis in the chick α7 subunit two distinct consensus Ca2+-binding sequences involved in this potentiation. It was suggested that they would be located on two calmodulin-like EF-hands that would form Ca2+-binding sites. However, in the model of α7, those residues localize along two different β-strands, ranging from the agonist-binding site, at an equatorial position, to the base of the amino-terminal domain. All the residues face outward and are located every two positions along the sequence from 161 to 169 (β-strand), except the residue E172. The two conclusions we can draw from that arrangement are, (i) the impossibility of an EF hand formed by identified residues close in the primary sequence (E161, D163, S165, Y167, S169, and E172), and (ii) the possible existence of several Ca2+-binding sites formed by these residues.
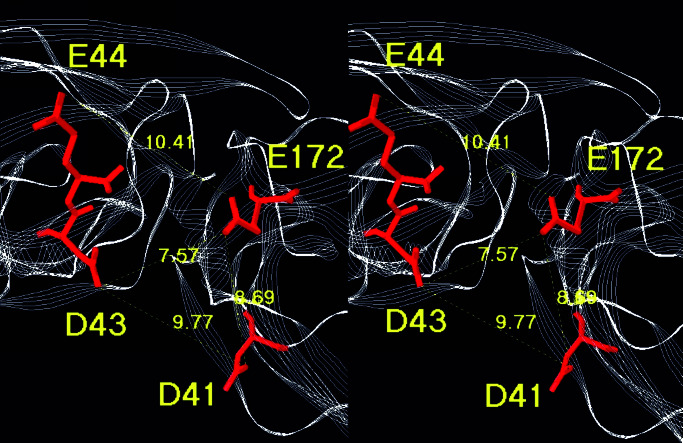
Among the mutated residues, E44 and E172 showed the strongest involvement. In particular, the mutation E172Q completely abolished the potentiation. The residues E44 and E172 of a given subunit are not only distant in the sequence, but they are also separated by 11.4 Å in the three-dimensional model (Fig. 5). In addition, they cannot physically interact within the subunit to form an intrasubunit Ca2+-binding site. In sharp contrast, residue E44 of the (+) subunit is closer to residue E172 of the (−) subunit (10.4 Å), and nothing precludes their involvement in the same intersubunit Ca2+-binding site. Even more interesting are residue D43 of the (+) subunit and residue D41 of the (−) subunit, not reported in ref. 58, which are closer to E172 (7.57 and 8.69 Å) and evenly located around a pocket that could accommodate a calcium ion. Thus, the chelated Ca2+ at subunit interfaces would favor certain relative positions of the adjacent subunits and therefore might affect the allosteric transitions, for instance by decreasing the allosteric constant BAL (58) and stabilizing the open state. In our model, the distances are too large for a strong calcium site, where the typical distances between chelating atoms range from 3 to 5 Å. However, the calcium site would reach its final conformation only in the open state, which could slightly differ from the putative desensitized state modeled here.
Figure 5.
Stereoscopic representation of the putative Ca2+-binding site at the interface of two subunits, viewed from the bottom of the amino-terminal domain.
D43 is not present on β2 (histidine), γ and β1 (asparagine), and δ (lysine) (Fig. 1). D41 is not present on β1 (isoleucine) and is replaced by a serine on β2, α1, γ, and δ. α7 pentamers would then possess five strong calcium-binding sites, comprising three acid residues. In contrast, the receptors (α4)2(β2)3 and (α1)2β1γδ would possess two sites with two acid residues and a serine, which in other systems have been shown to chelate calcium ions (for instance in synaptotagmin). This difference would explain the potentiation by calcium in (α7)5 (57) much stronger than in (α4)2(β2)3 (59) and (α1)2β1γδ (56).
We have presented here the modeling of the extracellular moiety of several nicotinic receptors, presumably in their high-affinity desensitized state. We showed that the agonist-binding site is globally conserved, although some specific positions should be crucial to pharmacological selectivity. Finally, the existence of an intersubunit chelating site for Ca2+ is proposed. These hypotheses should be testable by mutagenesis approaches. It is important to state that the models presented here are frozen “snapshots” of the receptor, constrained by the structure of the template we used, i.e., AChBP as presented in ref. 22. As a next step of our analysis, we plan to model the other conformations of the extracellular domain, basal and active, and also the transmembrane part of the nAChRs. Last, we plan to investigate the structure of the receptor in its dynamic dimension and to relate these data to the membrane physiology, in normal and pathologic situations.
Acknowledgments
We thank Andras Fiser for his help with modeller 6, Marc Delarue and Garrett Morris for their help in the docking process, Pierre-Jean Corringer, Stuart Edelstein, and Clément Léna for helpful discussions, and Matthew Levin for critical reading of the manuscript. The coordinates of AChBP were kindly provided by Drs. Titia Sixma and Katjuca Brejc. This work was supported by the Collège de France, the Commission of the European Communities (CEC), and the Association Française contre les Myopathies. T.G. is supported by CEC contract 2000-00318. All models presented in this paper can be retrieved from the Ligand-Gated Ion Channel database (http://www.pasteur.fr/recherche/banques/LGIC/LGIC.html).
Abbreviations
- nAChRs
nicotinic acetylcholine receptors
- AChBP
acetylcholine-binding protein
- rmsd
rms deviation
References
- 1.Changeux J-P, Edelstein S. Neuron. 1998;21:959–980. doi: 10.1016/s0896-6273(00)80616-9. [DOI] [PubMed] [Google Scholar]
- 2.Karlin A, Akabas M. Neuron. 1995;15:1231–1244. doi: 10.1016/0896-6273(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 3.Galzi J-L, Changeux J-P. Curr Opin Struct Biol. 1994;4:554–565. [Google Scholar]
- 4.Le Novère N, Changeux J-P. J Mol Evol. 1995;40:155–172. doi: 10.1007/BF00167110. [DOI] [PubMed] [Google Scholar]
- 5.Le Novère N, Changeux J-P. Nucleic Acids Res. 1999;27:340–342. doi: 10.1093/nar/27.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelstein S, Schaad O, Henry E, Bertrand D, Changeux J-P. Biol Cybernet. 1996;75:361–379. doi: 10.1007/s004220050302. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand D, Devillers-Thiéry A, Revah F, Galzi J-L, Hussy N, Mulle C, Bertrand S, Ballivet M, Changeux J-P. Proc Natl Acad Sci USA. 1992;89:1261–1265. doi: 10.1073/pnas.89.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore M, McCarthy M. Biochim Biophys Acta. 1995;1235:336–342. doi: 10.1016/0005-2736(95)80022-8. [DOI] [PubMed] [Google Scholar]
- 9.Middleton R, Strnad N, Cohen J. Mol Pharmacol. 1999;56:290–299. doi: 10.1124/mol.56.2.290. [DOI] [PubMed] [Google Scholar]
- 10.Fruchart-Gaillard C, Gilquin B, Antil-Delbeke S, Le Novère N, Tamiya T, Corringer P-J, Changeux J-P, Ménez A, Servent D. Proc Natl Acad Sci USA. 2002;99:3216–3221. doi: 10.1073/pnas.042699899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Changeux J-P, Kasai M, Lee C. Proc Natl Acad Sci USA. 1970;67:1241–1247. doi: 10.1073/pnas.67.3.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindstrom J. Mol Neurobiol. 1997;15:193–222. doi: 10.1007/BF02740634. [DOI] [PubMed] [Google Scholar]
- 13.Léna C, Changeux J-P. Curr Opin Neurobiol. 1997;7:674–682. doi: 10.1016/s0959-4388(97)80088-8. [DOI] [PubMed] [Google Scholar]
- 14.Arias A. Neurochem Int. 2000;36:595–645. doi: 10.1016/s0197-0186(99)00154-0. [DOI] [PubMed] [Google Scholar]
- 15.Corringer P-J, Le Novère N, Changeux J-P. Annu Rev Pharmacol Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 16.West A, Bjorkman P, Dougherty D, Lester H. J Biol Chem. 1997;272:25468–25473. doi: 10.1074/jbc.272.41.25468. [DOI] [PubMed] [Google Scholar]
- 17.Alexeev T, Krivoshein A, Shevalier A, Kudelina I, Telyakova O, Vincent A, Utkin Y, Hucho F, Tsetlin V. Eur J Biochem. 1999;259:310–319. doi: 10.1046/j.1432-1327.1999.00041.x. [DOI] [PubMed] [Google Scholar]
- 18.Schrattenholz A, Pfeiffer S, Pejovic V, Rudolph R, Godovac-Zimmermann J, Maelicke A. J Biol Chem. 1998;273:32393–32399. doi: 10.1074/jbc.273.49.32393. [DOI] [PubMed] [Google Scholar]
- 19.Wells G, Anand R, Wang F, Lindstrom J. J Biol Chem. 1998;273:964–973. doi: 10.1074/jbc.273.2.964. [DOI] [PubMed] [Google Scholar]
- 20.Le Novère N, Corringer P-J, Changeux J-P. Biophys J. 1999;76:2329–2345. doi: 10.1016/S0006-3495(99)77390-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unwin N. Philos Trans R Soc London B. 2000;355:1813–1829. doi: 10.1098/rstb.2000.0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brejc K, van Dijk W J, Klaassen R V, Schuurmans M, van der Oost J, Smit A B, Sixma T K. Nature (London) 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 23.Smit A B, Syed N I, Schaap D, van Minnen J, Klumperman J, Kits K S, Lodder H, van der Schors R C, van Elk R, Sorgedrager B, et al. Nature (London) 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 24.Russell R B, Saqi M A S, Sayle R A, Bates P A, Sternberg M J E. J Mol Biol. 1997;269:423–439. doi: 10.1006/jmbi.1997.1019. [DOI] [PubMed] [Google Scholar]
- 25.Rost B. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 26.Frishman D, Argos P. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 27.Guex N, Peitsch M. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sǎli A, Blundell T. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Thornton J. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris G M, Goodsell D S, Halliday R, Huey R, Hart W E, Belew R K, Olson A J. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- 32.Orengo C, Bray J, Hubbard T, LoConte L, Sillitoe I. Proteins. 1999;S3:149–170. doi: 10.1002/(sici)1097-0134(1999)37:3+<149::aid-prot20>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Paas Y, Devillers-Thiéry A, Teichberg V, Changeux J-P, Eisenstein M. Trends Pharmacol Sci. 2000;21:87–92. doi: 10.1016/s0165-6147(99)01443-1. [DOI] [PubMed] [Google Scholar]
- 34.Tiraboschi G, Jullian N, Thery V, Antonczak S, Fournie-Zaluski M, Roques B. Protein Eng. 1999;12:141–149. doi: 10.1093/protein/12.2.141. [DOI] [PubMed] [Google Scholar]
- 35.Martí-Renom M A, Stuart A C, Fiser A, Sánchez R, Melo F, Šali A. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 36.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 37.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 38.Green W, Wanamaker C. J Biol Chem. 1997;272:20945–20953. doi: 10.1074/jbc.272.33.20945. [DOI] [PubMed] [Google Scholar]
- 39.Cohen J, Sharp S D, Schyong Liu W. J Biol Chem. 1991;266:23354–23364. [PubMed] [Google Scholar]
- 40.Grutter T, Ehret-Sabatier L, Kotzyba-Hibert F, Goeldner M. Biochemistry. 2000;39:3034–3042. doi: 10.1021/bi992393o. [DOI] [PubMed] [Google Scholar]
- 41.Zhong W, Gallivan J P, Zhang Y, Li L, Lester H A, Dougherty D A. Proc Natl Acad Sci USA. 1998;95:12088–12093. doi: 10.1073/pnas.95.21.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anand R, Peng X, Lindstrom J. FEBS Lett. 1993;327:241–246. doi: 10.1016/0014-5793(93)80177-v. [DOI] [PubMed] [Google Scholar]
- 43.Quiram P A, Sine S M. J Biol Chem. 1998;273:11001–11006. doi: 10.1074/jbc.273.18.11001. [DOI] [PubMed] [Google Scholar]
- 44.Rao M, Olson A. Proteins. 1999;34:173–183. doi: 10.1002/(sici)1097-0134(19990201)34:2<173::aid-prot3>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 45.Grutter T, Changeux J-P. Trends Biochem Sci. 2001;26:459–463. doi: 10.1016/s0968-0004(01)01921-1. [DOI] [PubMed] [Google Scholar]
- 46.Harel M, Kasher R, Nicolas A, Guss J M, Balass M, Fridkin M, Smit A B, Brejc K, Sixma T K, Katchalski-Katzir E, et al. Neuron. 2001;32:265–275. doi: 10.1016/s0896-6273(01)00461-5. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen S, Cohen J. Proc Natl Acad Sci USA. 1990;87:2785–2789. doi: 10.1073/pnas.87.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez K, Corringer P, Edelstein S, Changeux J-P, Merola F. Biochemistry. 2000;39:6979–6990. doi: 10.1021/bi992811p. [DOI] [PubMed] [Google Scholar]
- 49.Sine S. Proc Natl Acad Sci USA. 1993;90:9436–9440. doi: 10.1073/pnas.90.20.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prince R, Sine S. J Biol Chem. 1998;273:7843–7849. doi: 10.1074/jbc.273.14.7843. [DOI] [PubMed] [Google Scholar]
- 51.Cohen B, Figl N, Quick M, Labarca C, Davidson N, Lester H. J Gen Physiol. 1995;105:745–764. doi: 10.1085/jgp.105.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker M J, Harvey S C, Luetje C W. J Pharmacol Exp Ther. 2001;299:385–391. [PubMed] [Google Scholar]
- 53.Léna C, Changeux J-P. Trends Neurosci. 1993;16:181–186. doi: 10.1016/0166-2236(93)90150-k. [DOI] [PubMed] [Google Scholar]
- 54.Mulle C, Léna C, Changeux J-P. Neuron. 1992;8:937–945. doi: 10.1016/0896-6273(92)90208-u. [DOI] [PubMed] [Google Scholar]
- 55.Vernino S, Amador M, Luetje C, Patrick J, Dani J. Neuron. 1992;8:127–134. doi: 10.1016/0896-6273(92)90114-s. [DOI] [PubMed] [Google Scholar]
- 56.Sine S, Claudio T, Sigworth F. J Gen Physiol. 1990;96:395–437. doi: 10.1085/jgp.96.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galzi J-L, Bertrand S, Corringer P-J, Changeux J-P, Bertrand D. EMBO J. 1996;15:5824–5832. [PMC free article] [PubMed] [Google Scholar]
- 58.Galzi J-L, Edelstein S, Changeux J-P. Proc Natl Acad Sci USA. 1996;93:1853–1858. doi: 10.1073/pnas.93.5.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buisson B, Gopalakrishnan M, Arneric S, Sullivan J, Bertrand D. J Neurosci. 1996;16:7880–7891. doi: 10.1523/JNEUROSCI.16-24-07880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]