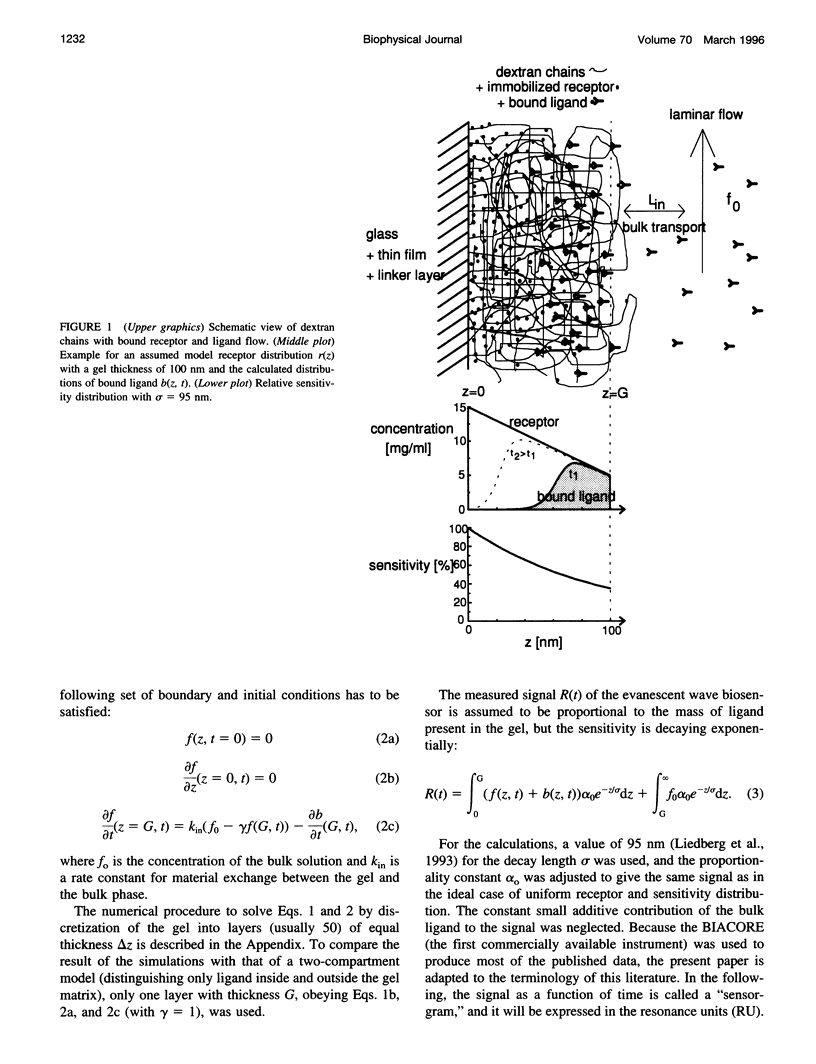
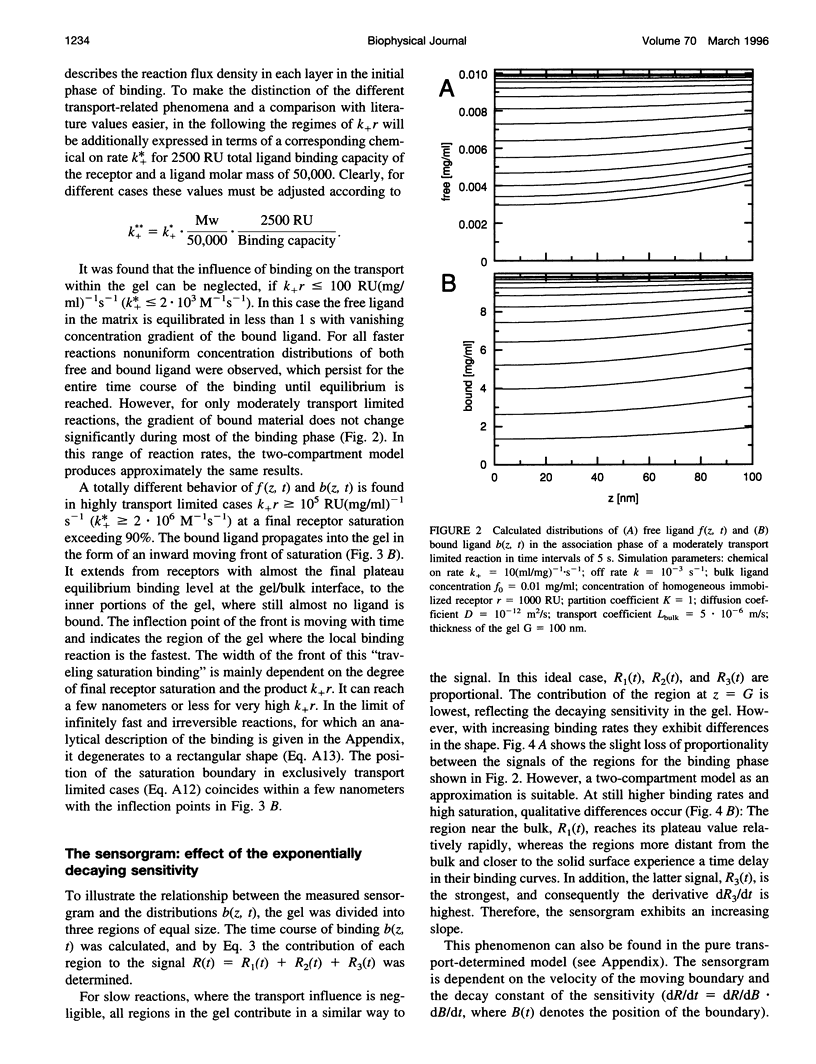
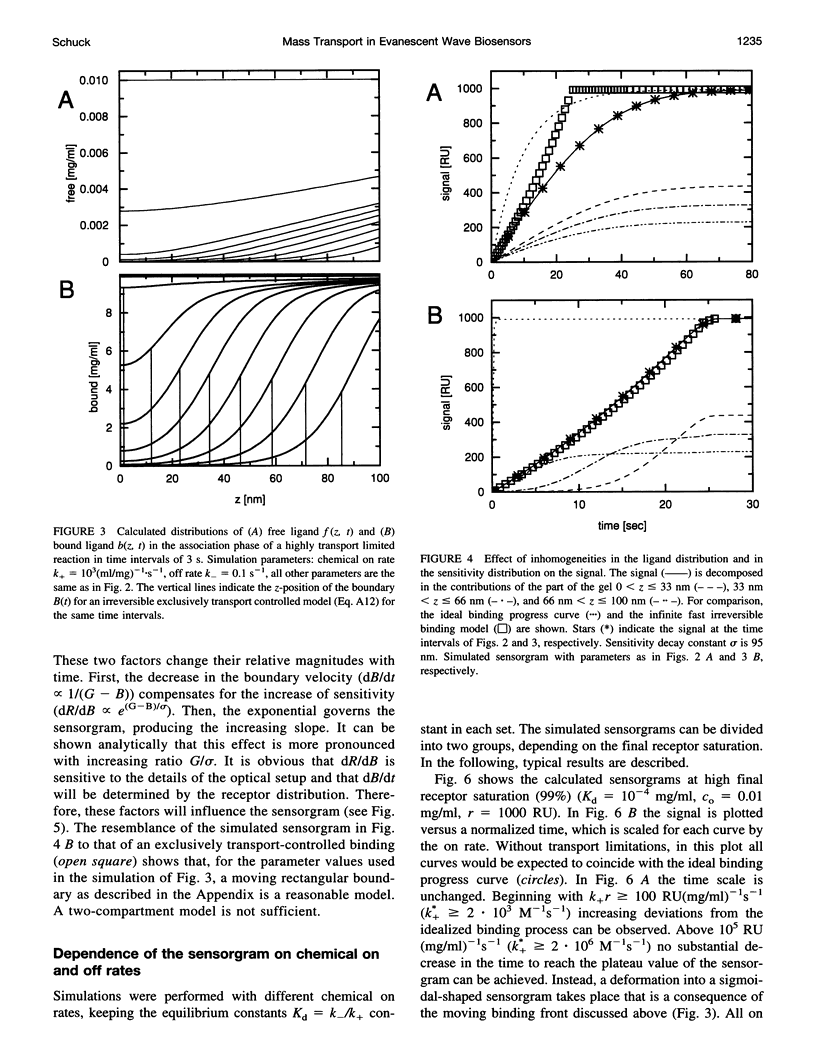
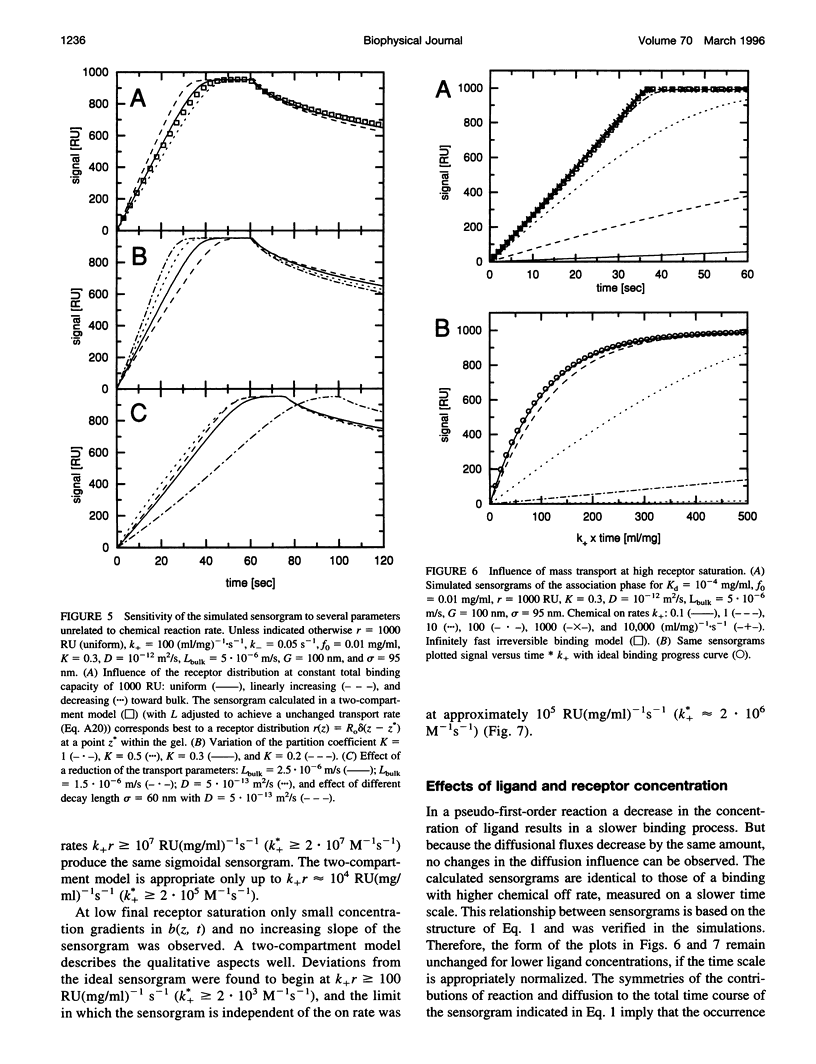
Abstract
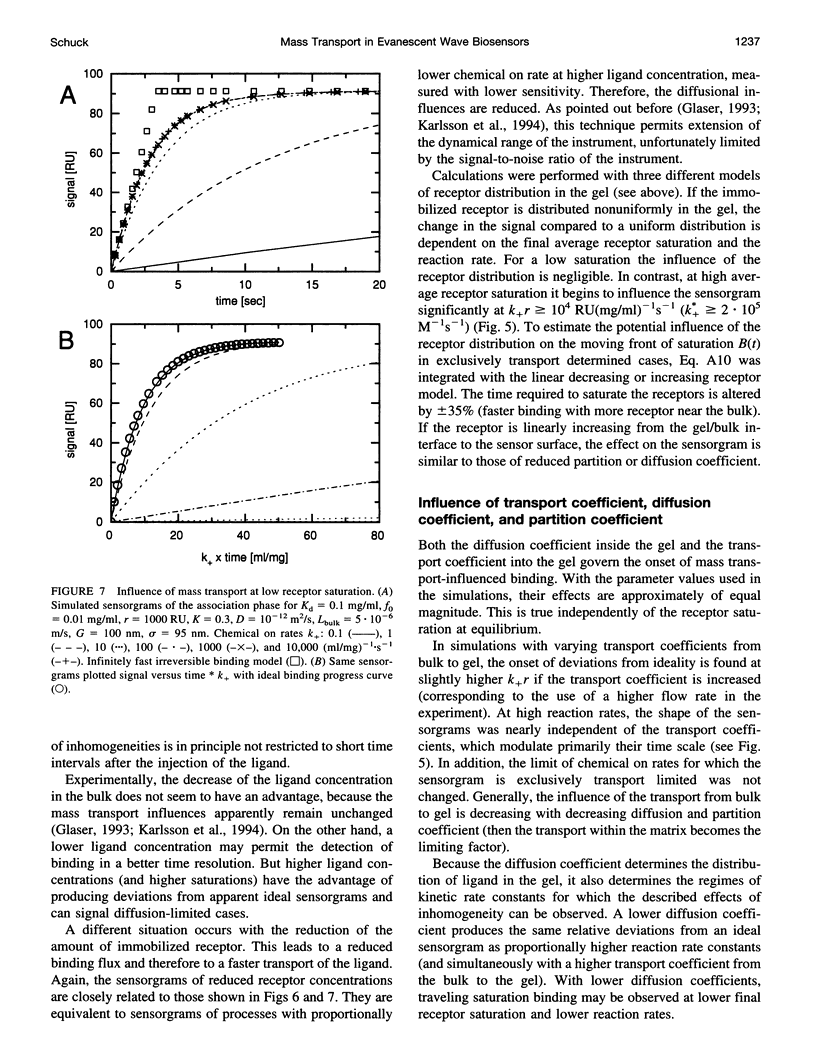
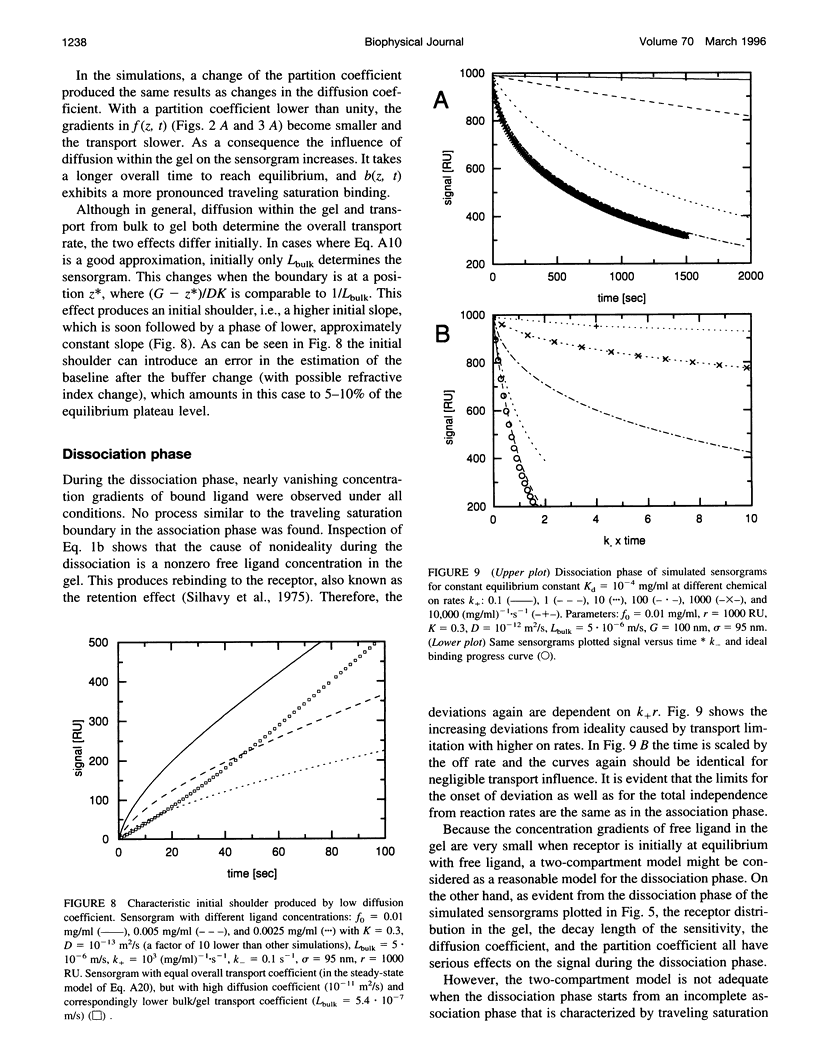
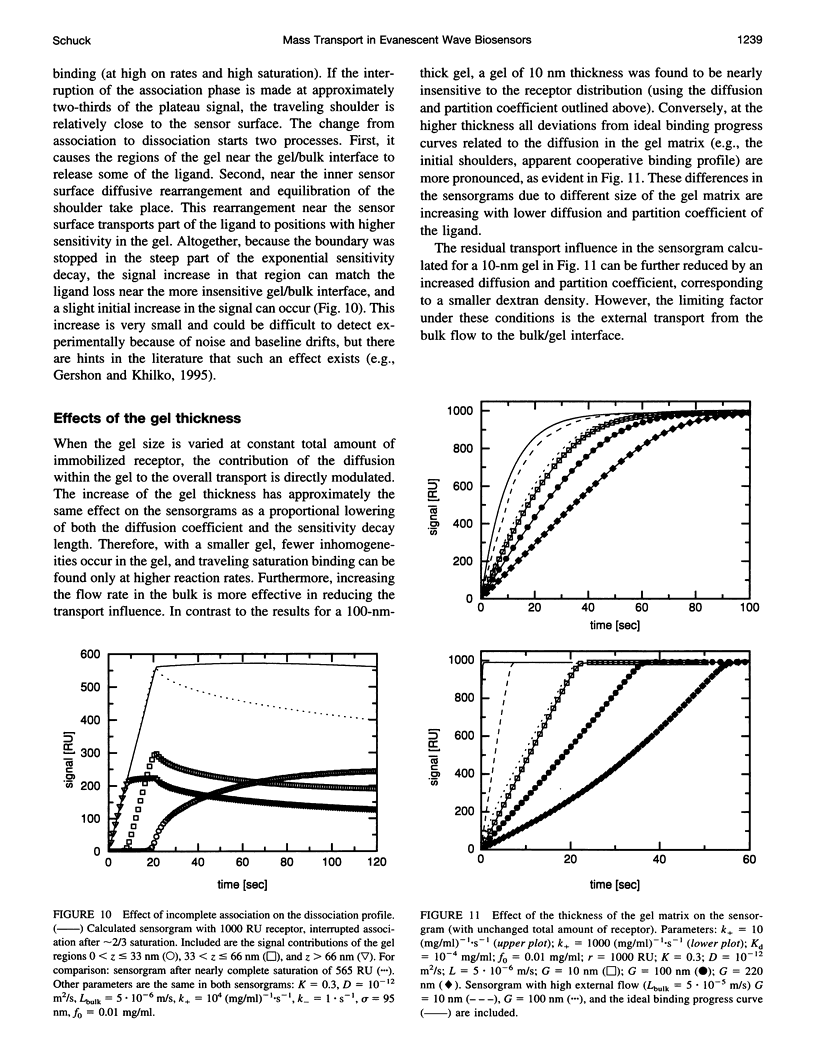
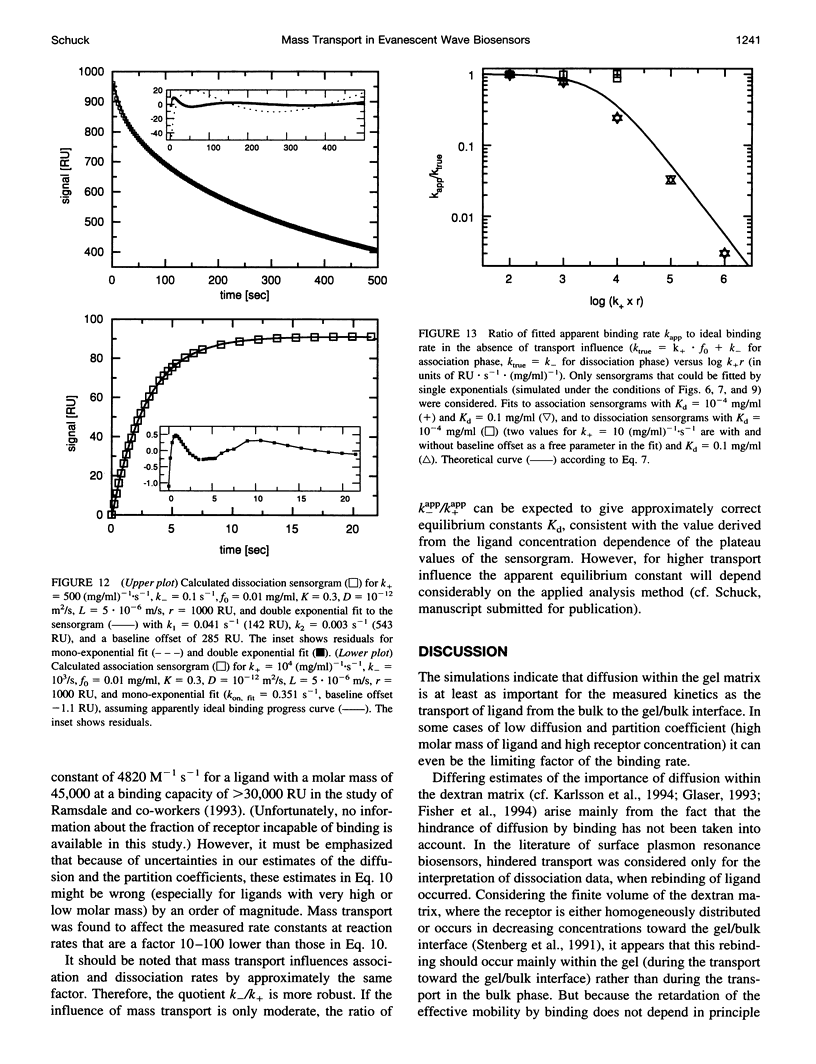
The influence of mass transport on ligand binding to receptor immobilized in a polymer matrix, as detected with an evanescent wave biosensor, was investigated. A one-dimensional computer model for the mass transport of ligand between the bulk solution and the polymer gel and within the gel was employed, and the influence of the diffusion coefficient, the partition coefficient, the thickness of the matrix, and the distribution of immobilized receptor were studied for a variety of conditions. Under conditions that may apply to many published experimental studies, diffusion within the matrix was found to decrease the overall ligand transport significantly. For relatively slow reactions, small spatial gradients of free and bound ligand in the gel are found, whereas for relatively rapid reactions strong inhomogeneities of ligand within the gel occur before establishment of equilibrium. Several types of deviations from ideal pseudo-first-order binding progress curves are described that resemble those of published experimental data. Extremely transport limited reactions can in some cases be fitted with apparently ideal binding progress curves, although with apparent reaction rates that are much lower than the true reaction rates. Nevertheless, the ratio of the apparent rate constants can be semiquantitatively consistent with the true equilibrium constant. Apparently "cooperative" binding can result from high chemical on rates at high receptor saturation. Dissociation in the presence of transport limitation was found to be well described empirically by a single or a double exponential, with both apparent rate constants considerably lower than the intrinsic chemical rate constant. Transport limitations in the gel can introduce many generally unknown factors into the binding progress curve. The simulations suggest that unexpected deviations from ideal binding progress curves may be due to highly transport influenced binding kinetics. The use of a thinner polymer matrix could significantly increase the range of detectable rate constants.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balgi G., Leckband D. E., Nitsche J. M. Transport effects on the kinetics of protein-surface binding. Biophys J. 1995 Jun;68(6):2251–2260. doi: 10.1016/S0006-3495(95)80407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrebaeck C. A., Malmborg A. C., Furebring C., Michaelsson A., Ward S., Danielsson L., Ohlin M. Kinetic analysis of recombinant antibody-antigen interactions: relation between structural domains and antigen binding. Biotechnology (N Y) 1992 Jun;10(6):697–698. doi: 10.1038/nbt0692-697. [DOI] [PubMed] [Google Scholar]
- Calakos N., Bennett M. K., Peterson K. E., Scheller R. H. Protein-protein interactions contributing to the specificity of intracellular vesicular trafficking. Science. 1994 Feb 25;263(5150):1146–1149. doi: 10.1126/science.8108733. [DOI] [PubMed] [Google Scholar]
- Chaiken I., Rosé S., Karlsson R. Analysis of macromolecular interactions using immobilized ligands. Anal Biochem. 1992 Mar;201(2):197–210. doi: 10.1016/0003-2697(92)90329-6. [DOI] [PubMed] [Google Scholar]
- Cooper L. J., Robertson D., Granzow R., Greenspan N. S. Variable domain-identical antibodies exhibit IgG subclass-related differences in affinity and kinetic constants as determined by surface plasmon resonance. Mol Immunol. 1994 Jun;31(8):577–584. doi: 10.1016/0161-5890(94)90165-1. [DOI] [PubMed] [Google Scholar]
- Corti A., Poiesi C., Merli S., Cassani G. Tumor necrosis factor (TNF) alpha quantification by ELISA and bioassay: effects of TNF alpha-soluble TNF receptor (p55) complex dissociation during assay incubations. J Immunol Methods. 1994 Dec 28;177(1-2):191–198. doi: 10.1016/0022-1759(94)90156-2. [DOI] [PubMed] [Google Scholar]
- DeLisi C. The biophysics of ligand-receptor interactions. Q Rev Biophys. 1980 May;13(2):201–230. doi: 10.1017/s0033583500001657. [DOI] [PubMed] [Google Scholar]
- Felder S., Zhou M., Hu P., Ureña J., Ullrich A., Chaudhuri M., White M., Shoelson S. E., Schlessinger J. SH2 domains exhibit high-affinity binding to tyrosine-phosphorylated peptides yet also exhibit rapid dissociation and exchange. Mol Cell Biol. 1993 Mar;13(3):1449–1455. doi: 10.1128/mcb.13.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. J., Fivash M. Surface plasmon resonance based methods for measuring the kinetics and binding affinities of biomolecular interactions. Curr Opin Biotechnol. 1994 Aug;5(4):389–395. doi: 10.1016/0958-1669(94)90047-7. [DOI] [PubMed] [Google Scholar]
- Foote J., Milstein C. Conformational isomerism and the diversity of antibodies. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote J., Milstein C. Kinetic maturation of an immune response. Nature. 1991 Aug 8;352(6335):530–532. doi: 10.1038/352530a0. [DOI] [PubMed] [Google Scholar]
- Fägerstam L. G., Frostell-Karlsson A., Karlsson R., Persson B., Rönnberg I. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J Chromatogr. 1992 Apr 24;597(1-2):397–410. doi: 10.1016/0021-9673(92)80137-j. [DOI] [PubMed] [Google Scholar]
- George A. J., French R. R., Glennie M. J. Measurement of kinetic binding constants of a panel of anti-saporin antibodies using a resonant mirror biosensor. J Immunol Methods. 1995 Jun 14;183(1):51–63. doi: 10.1016/0022-1759(95)00031-5. [DOI] [PubMed] [Google Scholar]
- Gershon P. D., Khilko S. Stable chelating linkage for reversible immobilization of oligohistidine tagged proteins in the BIAcore surface plasmon resonance detector. J Immunol Methods. 1995 Jun 14;183(1):65–76. doi: 10.1016/0022-1759(95)00032-6. [DOI] [PubMed] [Google Scholar]
- Glaser R. W. Antigen-antibody binding and mass transport by convection and diffusion to a surface: a two-dimensional computer model of binding and dissociation kinetics. Anal Biochem. 1993 Aug 15;213(1):152–161. doi: 10.1006/abio.1993.1399. [DOI] [PubMed] [Google Scholar]
- Goldstein B., Dembo M. Approximating the effects of diffusion on reversible reactions at the cell surface: ligand-receptor kinetics. Biophys J. 1995 Apr;68(4):1222–1230. doi: 10.1016/S0006-3495(95)80298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard G., Benkirane N., Zeder-Lutz G., van Regenmortel M. H., Briand J. P., Muller S. Antigenic mimicry of natural L-peptides with retro-inverso-peptidomimetics. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9765–9769. doi: 10.1073/pnas.91.21.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschemeyer R. H., Bowers W. F. Exponential analysis of concentration or concentration difference data for discrete molecular weight distributions in sedimentation equilibrium. Biochemistry. 1970 Jan 20;9(2):435–445. doi: 10.1021/bi00804a035. [DOI] [PubMed] [Google Scholar]
- Horenstein A. L., Poiesi C., DeMonte L., Camagna M., Mariani M., Albertini A., Malavasi F. Real-time kinetic analysis applied to the production of bispecific monoclonal antibodies for radioimmunodetection of cancer. Int J Clin Lab Res. 1993;23(4):199–205. doi: 10.1007/BF02592309. [DOI] [PubMed] [Google Scholar]
- Hsieh H. V., Thompson N. L. Theory for measuring bivalent surface binding kinetics using total internal reflection with fluorescence photobleaching recovery. Biophys J. 1994 Mar;66(3 Pt 1):898–911. doi: 10.1016/s0006-3495(94)80866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito W., Kurosawa Y. Development of an artificial antibody system with multiple valency using an Fv fragment fused to a fragment of protein A. J Biol Chem. 1993 Sep 25;268(27):20668–20675. [PubMed] [Google Scholar]
- Johne B., Gadnell M., Hansen K. Epitope mapping and binding kinetics of monoclonal antibodies studied by real time biospecific interaction analysis using surface plasmon resonance. J Immunol Methods. 1993 Apr 2;160(2):191–198. doi: 10.1016/0022-1759(93)90177-9. [DOI] [PubMed] [Google Scholar]
- Johnsson B., Löfås S., Lindquist G., Edström A., Müller Hillgren R. M., Hansson A. Comparison of methods for immobilization to carboxymethyl dextran sensor surfaces by analysis of the specific activity of monoclonal antibodies. J Mol Recognit. 1995 Jan-Apr;8(1-2):125–131. doi: 10.1002/jmr.300080122. [DOI] [PubMed] [Google Scholar]
- Jönsson U., Fägerstam L., Ivarsson B., Johnsson B., Karlsson R., Lundh K., Löfås S., Persson B., Roos H., Rönnberg I. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. Biotechniques. 1991 Nov;11(5):620–627. [PubMed] [Google Scholar]
- Jönsson U., Fägerstam L., Löfas S., Stenberg E., Karlsson R., Frostell A., Markey F., Schindler F. Introducing a biosensor based technology for real-time biospecific interaction analysis. Ann Biol Clin (Paris) 1993;51(1):19–26. [PubMed] [Google Scholar]
- Karlsson R., Michaelsson A., Mattsson L. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991 Dec 15;145(1-2):229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- Malmborg A. C., Borrebaeck C. A. BIAcore as a tool in antibody engineering. J Immunol Methods. 1995 Jun 14;183(1):7–13. doi: 10.1016/0022-1759(95)00018-6. [DOI] [PubMed] [Google Scholar]
- Malmborg A. C., Michaëlsson A., Ohlin M., Jansson B., Borrebaeck C. A. Real time analysis of antibody-antigen reaction kinetics. Scand J Immunol. 1992 Jun;35(6):643–650. doi: 10.1111/j.1365-3083.1992.tb02970.x. [DOI] [PubMed] [Google Scholar]
- Mani J. C., Marchi V., Cucurou C. Effect of HIV-1 peptide presentation on the affinity constants of two monoclonal antibodies determined by BIAcore technology. Mol Immunol. 1994 Apr;31(6):439–444. doi: 10.1016/0161-5890(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Morton T. A., Bennett D. B., Appelbaum E. R., Cusimano D. M., Johanson K. O., Matico R. E., Young P. R., Doyle M., Chaiken I. M. Analysis of the interaction between human interleukin-5 and the soluble domain of its receptor using a surface plasmon resonance biosensor. J Mol Recognit. 1994 Mar;7(1):47–55. doi: 10.1002/jmr.300070107. [DOI] [PubMed] [Google Scholar]
- Natsume T., Koide T., Yokota S., Hirayoshi K., Nagata K. Interactions between collagen-binding stress protein HSP47 and collagen. Analysis of kinetic parameters by surface plasmon resonance biosensor. J Biol Chem. 1994 Dec 9;269(49):31224–31228. [PubMed] [Google Scholar]
- O'Shannessy D. J., Brigham-Burke M., Soneson K. K., Hensley P., Brooks I. Determination of rate and equilibrium binding constants for macromolecular interactions using surface plasmon resonance: use of nonlinear least squares analysis methods. Anal Biochem. 1993 Aug 1;212(2):457–468. doi: 10.1006/abio.1993.1355. [DOI] [PubMed] [Google Scholar]
- O'Shannessy D. J. Determination of kinetic rate and equilibrium binding constants for macromolecular interactions: a critique of the surface plasmon resonance literature. Curr Opin Biotechnol. 1994 Feb;5(1):65–71. doi: 10.1016/s0958-1669(05)80072-2. [DOI] [PubMed] [Google Scholar]
- Pack P., Müller K., Zahn R., Plückthun A. Tetravalent miniantibodies with high avidity assembling in Escherichia coli. J Mol Biol. 1995 Feb 10;246(1):28–34. doi: 10.1006/jmbi.1994.0062. [DOI] [PubMed] [Google Scholar]
- Panayotou G., Gish G., End P., Truong O., Gout I., Dhand R., Fry M. J., Hiles I., Pawson T., Waterfield M. D. Interactions between SH2 domains and tyrosine-phosphorylated platelet-derived growth factor beta-receptor sequences: analysis of kinetic parameters by a novel biosensor-based approach. Mol Cell Biol. 1993 Jun;13(6):3567–3576. doi: 10.1128/mcb.13.6.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons I. D., Persson B., Mekhalfia A., Blackburn G. M., Stockley P. G. Probing the molecular mechanism of action of co-repressor in the E. coli methionine repressor-operator complex using surface plasmon resonance (SPR). Nucleic Acids Res. 1995 Jan 25;23(2):211–216. doi: 10.1093/nar/23.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G., Shoelson S. E., Gish G. D., Pawson T., Walsh C. T. Kinetics of p56lck and p60src Src homology 2 domain binding to tyrosine-phosphorylated peptides determined by a competition assay or surface plasmon resonance. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):4902–4906. doi: 10.1073/pnas.90.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillies G. D. Diffusion of bovine serum albumin in a neutral polymer solution. Biopolymers. 1985 Feb;24(2):379–386. doi: 10.1002/bip.360240206. [DOI] [PubMed] [Google Scholar]
- Plant A. L., Brigham-Burke M., Petrella E. C., O'Shannessy D. J. Phospholipid/alkanethiol bilayers for cell-surface receptor studies by surface plasmon resonance. Anal Biochem. 1995 Apr 10;226(2):342–348. doi: 10.1006/abio.1995.1234. [DOI] [PubMed] [Google Scholar]
- Poiesi C., Albertini A., Ghielmi S., Cassani G., Corti A. Kinetic analysis of TNF-alpha oligomer-monomer transition by surface plasmon resonance and immunochemical methods. Cytokine. 1993 Nov;5(6):539–545. doi: 10.1016/s1043-4666(05)80002-x. [DOI] [PubMed] [Google Scholar]
- Ramsdale T. E., Andrews P. R., Nice E. C. Verification of the interaction between peptide T and CD4 using surface plasmon resonance. FEBS Lett. 1993 Nov 1;333(3):217–222. doi: 10.1016/0014-5793(93)80657-g. [DOI] [PubMed] [Google Scholar]
- Richalet-Sécordel P. M., Zeder-Lutz G., Plaue S., Sommermeyer-Leroux G., Van Regenmortel M. H. Cross-reactivity of monoclonal antibodies to a chimeric V3 peptide of HIV-1 with peptide analogues studied by biosensor technology and ELISA. J Immunol Methods. 1994 Dec 2;176(2):221–234. doi: 10.1016/0022-1759(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Szmelcman S., Boos W., Schwartz M. On the significance of the retention of ligand by protein. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2120–2124. doi: 10.1073/pnas.72.6.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjölander S., Urbaniczky C. Integrated fluid handling system for biomolecular interaction analysis. Anal Chem. 1991 Oct 15;63(20):2338–2345. doi: 10.1021/ac00020a025. [DOI] [PubMed] [Google Scholar]
- Takano E., Hatanaka M., Maki M. Real-time-analysis of the calcium-dependent interaction between calmodulin and a synthetic oligopeptide of calcineurin by a surface plasmon resonance biosensor. FEBS Lett. 1994 Sep 26;352(2):247–250. doi: 10.1016/0014-5793(94)00965-1. [DOI] [PubMed] [Google Scholar]
- Takano E., Ma H., Yang H. Q., Maki M., Hatanaka M. Preference of calcium-dependent interactions between calmodulin-like domains of calpain and calpastatin subdomains. FEBS Lett. 1995 Mar 27;362(1):93–97. doi: 10.1016/0014-5793(95)00219-y. [DOI] [PubMed] [Google Scholar]
- Tosser G., Delaunay T., Kohli E., Grosclaude J., Pothier P., Cohen J. Topology of bovine rotavirus (RF strain) VP6 epitopes by real-time biospecific interaction analysis. Virology. 1994 Oct;204(1):8–16. doi: 10.1006/viro.1994.1505. [DOI] [PubMed] [Google Scholar]
- Watts H. J., Lowe C. R., Pollard-Knight D. V. Optical biosensor for monitoring microbial cells. Anal Chem. 1994 Aug 1;66(15):2465–2470. doi: 10.1021/ac00087a010. [DOI] [PubMed] [Google Scholar]
- Wohlhueter R. M., Parekh K., Udhayakumar V., Fang S., Lal A. A. Analysis of binding of monoclonal antibody to a malarial peptide by surface plasmon resonance biosensor and integrated rate equations. J Immunol. 1994 Jul 1;153(1):181–189. [PubMed] [Google Scholar]
- Yang H. Q., Ma H., Takano E., Hatanaka M., Maki M. Analysis of calcium-dependent interaction between amino-terminal conserved region of calpastatin functional domain and calmodulin-like domain of mu-calpain large subunit. J Biol Chem. 1994 Jul 22;269(29):18977–18984. [PubMed] [Google Scholar]
- Zahn R., Axmann S. E., Rücknagel K. P., Jaeger E., Laminet A. A., Plückthun A. Thermodynamic partitioning model for hydrophobic binding of polypeptides by GroEL. I. GroEL recognizes the signal sequences of beta-lactamase precursor. J Mol Biol. 1994 Sep 16;242(2):150–164. doi: 10.1006/jmbi.1994.1566. [DOI] [PubMed] [Google Scholar]
- Zwanzig R., Szabo A. Time dependent rate of diffusion-influenced ligand binding to receptors on cell surfaces. Biophys J. 1991 Sep;60(3):671–678. doi: 10.1016/S0006-3495(91)82096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe P. A., Brown M. H., Davis S. J., Barclay A. N. Affinity and kinetic analysis of the interaction of the cell adhesion molecules rat CD2 and CD48. EMBO J. 1993 Dec 15;12(13):4945–4954. doi: 10.1002/j.1460-2075.1993.tb06188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


