Abstract
Parkinson's disease is a complex neurodegenerative disorder characterized by the death of brain dopamine neurons. In mammals, dopamine neuronal degeneration can be triggered through exposure to neurotoxins accumulated by the presynaptic dopamine transporter (DAT), including 6-hydroxydopamine (6-OHDA) and 1-methyl-4-phenylpyridinium. We have established a system for the pharmacological and genetic evaluation of neurotoxin-induced dopamine neuronal death in Caenorhabditis elegans. Brief (1 h) exposure of green fluorescent protein-tagged, living worms to 6-OHDA causes selective degeneration of dopamine neurons. We demonstrate that agents that interfere with DAT function protect against 6-OHDA toxicity. 6-OHDA-triggered neural degeneration does not require the CED-3/CED-4 cell death pathway, but is abolished by the genetic disruption of the C. elegans DAT.
Keywords: transporter‖genetics‖catecholamine‖Parkinson's disease‖apoptosis
Parkinson's disease (PD), a severe movement disorder characterized by resting tremor, spasticity, and an inability to initiate movement, arises from the progressive, irreversible loss of dopamine neurons in the substantia nigra pars compacta (1). Although rare genetic forms of PD have been identified (2, 3), the molecular determinants of dopamine neuron vulnerability and cell death in the majority of PD cases remain as yet ill defined. Current hypotheses involve environmental toxin exposure, the increased generation of reactive oxygen species, and inhibition of mitochondrial electron transport (1, 4). How these events are targeted to dopamine neurons is unresolved. Dopamine itself is a highly oxidizable substance, and its uncontrolled metabolism or improper sequestration may trigger oxidative damage, supporting cell death (5, 6).
The best-studied rodent and primate models of PD use the neurotoxins 6-hydroxydopamine (6-OHDA) or 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), and more recently the pesticide rotenone, to chemically ablate dopamine neurons in vivo (1, 7, 8). 6-OHDA and the reactive metabolite of MPTP, MPP+ (1-methyl-4-phenylpyridinium), are selectively accumulated by dopamine neurons through preynaptic dopamine transporters (DATs), causing an increase in reactive oxygen species and/or mitochondrial dysfunction, and that may thereby induce neuritic damage and cell death (9–11). 6-OHDA is particularly intriguing as it is found in brain and urine samples of PD patients, suggesting it may be an endogenous component of PD pathogenesis (12–14). Although mammalian models afford the highest degree of physiologic and behavioral correlates to PD, genetic manipulations that might define molecular pathways supporting dopamine neuron vulnerability, death, and protection are limited and often impractical. We reasoned that the presence of dopamine neurons in the nematode Caenorhabditis elegans (15), a transparent organism with well-developed genetic and transgenic methodologies, might afford new opportunities to investigate the molecular basis of dopamine neuron degeneration. At a molecular level, all of the fundamental components involved in dopamine neurotransmission are present in the nematode, including the proteins responsible for dopamine biosynthesis, vesicular packaging, and inactivation (16–19). Moreover, the genetics and cellular features of necrotic and apoptotic cell death have been well described in C. elegans (20, 21), affording an opportunity to relate toxin-induced dopamine neuronal injury to these pathways.
Materials and Methods
Strains and Maintenance.
C. elegans strains were cultured on bacterial lawns of either OP-50 or NA-22 at 22°C according to standard methods (22). N2 Bristol is the wild-type strain. ced-1(e1735), ced-3(n717), ced-4(n1162), dpy-20(e1282), dpy-17(e164), unc-13(e51), unc-30(e191), and unc79(e1068) strains were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis). The dat-1(ok157) strain was a gift of J. Duerr and J. Rand (Oklahoma Medical Research Foundation, Oklahoma City). The daf-7∷gfp line (DR2022) was a gift from D. Riddle (University of Missouri, Columbia) (23). The tph-1∷gfp (GR1321) was a gift of J. Sze (University of California, Irvine) and G. Ruvkun (Massachusetts General Hospital, Boston). All 6-OHDA assays involving ced mutants were performed in genetic backgrounds containing closely linked recessive markers to aid in identifying homozygous ced mutants: ced-1(e1735) unc-13(e51) (MT3608), ced-3(n717) unc-30(e191) dpy-20(e1282) (MT5729), and ced-4(n1162) unc-79(e1068) (MT2550). 6-OHDA experiments involving ced-4(n1162) dpy-17(e164) (MT2551) give qualitatively similar results as ced-4(n1162) unc-79(e1068). ced-3(n717) and ced-4(n1162) are likely null and/or strong loss-of-function alleles, respectively (24–26). ced-3 and ced-4 mutant lines show supernumerary dopamine neurons, further confirming the homozygosity of the ced mutations (data not shown).
Plasmid Construction.
The DAT-1 transcriptional green fluorescent protein (GFP) fusion (Pdat-1∷GFP) was created by PCR amplification of the DAT-1 regulatory region found in cosmid T23G5.5 (refs. 27 and 28; T. Ishihara and I. Katsura, personal communication). The amplified product, containing 716 bp immediately upstream of the initiating codon ATG, was digested with HindIII and BamHI, and the resultant 0.7-kb fragment was cloned into the HindIII and BamHI sites of pPD95.73 (gift from A. Fire, Carnigie Institute of Washington, Baltimore), now called pRB490. The DAT-1 translational GFP fusion (Pdat-1∷DAT-1∷GFP) was created by PCR amplification from cosmid T23G5.5 by using the above promoter to +0.4 kb from the initiating codon. The amplified product, which contained the first exon and intron and part of exon 2, was digested with HindIII and SpHI, and the resultant 1.2-kb fragment was cloned into the HindIII and SpHI sites of pPD95.73l (gift from A. Fire), now called pRB491. The remainder of the DAT-1 coding region was amplified by PCR at +1.1 kb from the initiation codon to immediately before the TGA stop codon by using the DAT-1 vector pRB235 [previously called pCeDAT2 (16)], as the template. The PCR fragment was digested with SpHI and BamHI, and the resultant 1.8-kb fragment was ligated in-frame into the same sites in pRB491, now called pRB493. For mammalian cell expression studies, DAT-1 cDNA was subcloned from pCeDAT2 (16) into pcDNA3 (Invitrogen) at KpnI and NotI sites, creating pRB454.
COS-7 Expression of DAT-1.
Functional expression of DAT-1 cDNA was obtained in transiently transfected COS cells by using the Fugene 6 Transfection reagent (Roche Diagnostics) following the manufacturer's protocol. Briefly, COS cells were cultured in DMEM supplemented with 10% FBS (HyClone), 2 mM l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. One day before transfections, 1 × 105 cells were plated in 24-well tissue culture plates. Cells were transfected with pRB454, and 48 h later cells were harvested and processed for [3H]dopamine transport as described (16), using [3H]dopamine at 50 nM, assaying 5 min at 37°C. Nonspecific transport, defined as accumulation in vector-transfected cells, was subtracted from the data to yield specific uptake. Inhibitory potency was determined by using nonlinear fits of transport data with GraphPad (San Diego) prism software.
Germ-Line Transformation.
Transgenic animals containing the transcriptional fusion (pRB490) were obtained after coinjection of 20 ng/ul of pRB490 Pdat-1∷GFP, 30 ng/ul of plasmid carrier DNA (pBluescript), and 50 ng/ul of pRF4 [rol-6(su1006)] into the N2 strain (29, 30). Animals were cultured at 24°C, and transgenic animals were selected by their Roller phenotype. Two independent integrants of the pRB490 array were obtained by gamma irradiation and both displayed similar GFP expression patterns; one integrant, BY200, was outcrossed four times and used for subsequent analysis. The BY200 insert was mapped to within 2.4 map units of dpy-11 on chromosome V. Transgenic animals containing the DAT-1 translational fusion (pRB493) also were generated by coselection with the rol-6 dominant marker, pRF4.
Exposure of C. elegans to 6-OHDA.
Embryos were obtained by hypochlorite treatment of gravid adults (31). After 17–24 h incubation in M9 buffer to obtain synchronized L1s, the worms were washed once in 10 ml dH2O, spread on 8P/NA22 plates, and incubated at 24°C for 25–29 h. L3 larvae were collected, washed in dH2O, and added to the assay mix (to a final OD600 worms of 0.1–0.2) with 2 or 10 mM ascorbic acid (AA) and 10 or 50 mM 6-hydroxydopamine ± 10 mM d-amphetamine or 1 mM imipramine. The assay solution (1–2 ml) was incubated for 1 h at 24°C and mixed gently every 15 min. The worms were then washed in dH2O and spread on NGM/OP50 plates. Worms immobilized with 2% NaN3 were scored 40–80 h post-6-OHDA exposure on 2% agarose pads.
For quantitative analyses of 6-OHDA-induced changes in cell morphology, 50–60 worms were applied to an agar pad and examined under a Kramer/Zeiss M2 fluorescent dissecting scope with a Plan Apo 10×/0.28 LWD objective (32 mm) or an inverted Axiovert S100 with a Plan Neofluar 40×/0.75 objective. For dopamine neuron analyses using the DAT-1 promotor, all four cephalic sensilla (CEP) dendrites were examined, and GFP fluorescence within the dendrites followed from the nerve ring to the tip of the nose. If any part of the dendrite was absent, the worm was considered to have altered CEPs and was scored as positive for dopamine neurodegeneration. For dopamine neurons analyses using the cat-2 promoter, the neurons were considered positive for degeneration if the any of the four CEP dendritic endings in the nose were absent. For the tph-1∷gfp-expressing cells, the neurons were scored as positive if either of the NSM cell bodies were damaged or missing. For the unc-4∷gfp-expressing cells, the neurons were scored as positive if anteriorly directed axons from the retrovesicular ganglion motor neurons, SABVL and SABVR, were absent. For the daf-7∷gfp-expressing cells, the neurons were scored as positive if the ASI cell bodies or processes could not be seen.
Microscopy.
Confocal images were captured with either a Zeiss LSM 410 or 510 confocal microscope. Z-series images were collected by optical sectioning at intervals of ≈1 μm. Epifluorescence micrographs were taken with Elite 400 film (Kodak) and a 35-mm camera mounted on a Zeiss Axioplan microscope with a ×63 or ×40 Plan Apo, N.A. 1.4 objective. All image processing and montage assembly were performed with Adobe photoshop.
For electron microscopy (EM), treated animals were fixed at ≈60 h after exposure to 6-OHDA, as described above. Primary fixation was in buffered aldehydes (2.5% glutaraldehyde plus 1.0% paraformaldehyde), followed by secondary fixation in 1% osmium tetroxide plus 0.5% potassium ferrocyanide, and en bloc staining with 1% uranyl acetate (UAc) (20). Samples were encased in 3% agarose, dehydrated, and embedded in Epon 812 resin (Electron Microscopy Sciences, Fort Washington, PA). Transverse serial thin sections through the head were collected on a diamond knife and poststained with UAc again and lead citrate. Dopamine neurons were examined on a Philips CM10 electron microscope.
Results
Visualization of C. elegans Dopamine Neurons and Sensitivity to 6-OHDA.
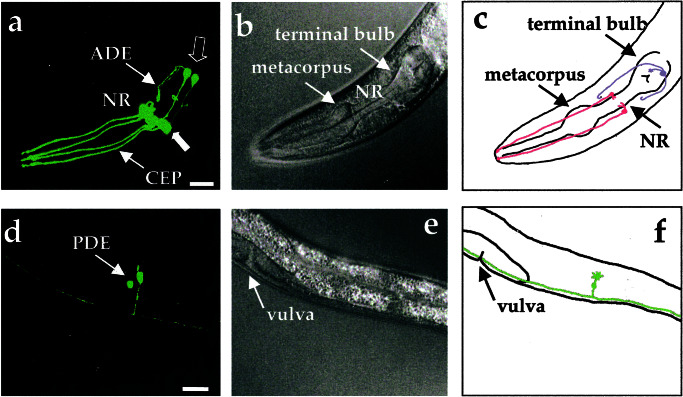
The C. elegans hermaphrodite possesses four bilaterally symmetric pairs of dopamine neurons that include three pairs within the head [two pairs of CEP neurons and one pair of anterior deirids (ADE) neurons] and one pair that is located in a posterior lateral position [posterior deirids (PDE) neurons] (15, 32). These neurons contain ciliated dendrites that contact the cuticle and likely convey mechanosensory information. Laser ablation and genetic studies implicate them in foraging, movement, and egg-laying behaviors (33–35). Previous studies using β-galactosidase fusions indicate that the cell bodies of these neurons can be visualized in fixed nematodes by using DAT (TG235.5; DAT-1) promoter elements to confer cell-specific expression (refs. 27 and 28; T. Ishihara and I. Katsura, personal communication). To visualize dopamine neurons and neuritic processes in living animals, we produced integrated transgenic lines driving GFP expression with ≈0.7 kb of the 5′ flanking sequence of DAT-1 (Pdat-1∷GFP; BY200). We observed intense GFP expression in all three classes of dopamine neurons in the hermaphrodite without any apparent expression in other cell types (Fig 1 a and d; see Movie 1, which is available as supporting information on the PNAS web site, www.pnas.org). Labeled cells match the position of cells visualized with tyrosine hydroxylase (cat-2) GFP reporter construct (EM641, gift of R. Lints and S. Emmons, Albert Einstein College of Medicine; data not shown). GFP expression was first evident in embryos, whereas expression within the posterior neurons was observed only postembryonically, consistent with the known ontogeny of the PDEs (36). In addition, the three pairs of tail dopamine neurons can be visualized in males derived from our Pdat-1∷GFP line (R. Lints and S. Emmons, personal communication), supporting a restricted expression of the DAT-1 promoter to dopaminergic neurons.
Figure 1.
Visualization of all eight dopamine neurons in living, adult C. elegans hermaphrodites by using DAT-1∷GFP transcriptional fusions. (a) Three-dimensional reconstruction of confocal epifluorescence from head dopamine neurons in a Pdat-1∷GFP transgenic line (BY200). Thin arrows identify CEP and ADE processes. Thick closed arrow points to four CEP cell bodies. Thick open arrow indicates location of two ADE cell bodies. NR refers to the nerve ring. (b) Differential interference contrast microscopy image of animal in a. (c) Schematic drawing showing location of dopamine neurons in the head relative to the pharynx. In this top view two pairs of CEP neurons (red) project dendritic endings to the tip of the nose and one pair of ADE neurons (blue) extend ciliated processes to amphids adjacent to the terminal bulb of the pharynx. (d) Three-dimensional reconstructions of confocal epifluorescence of the PDE neurons. Both PDE cell bodies are apparent. (e) Differential interference contrast microscopy image of animal in d. (f) Schematic drawing showing left-hand member of pair of PDE neurons (green) in lateral location posterior to vulva. (All scale bars = 25 μm.) Anterior is to the left.
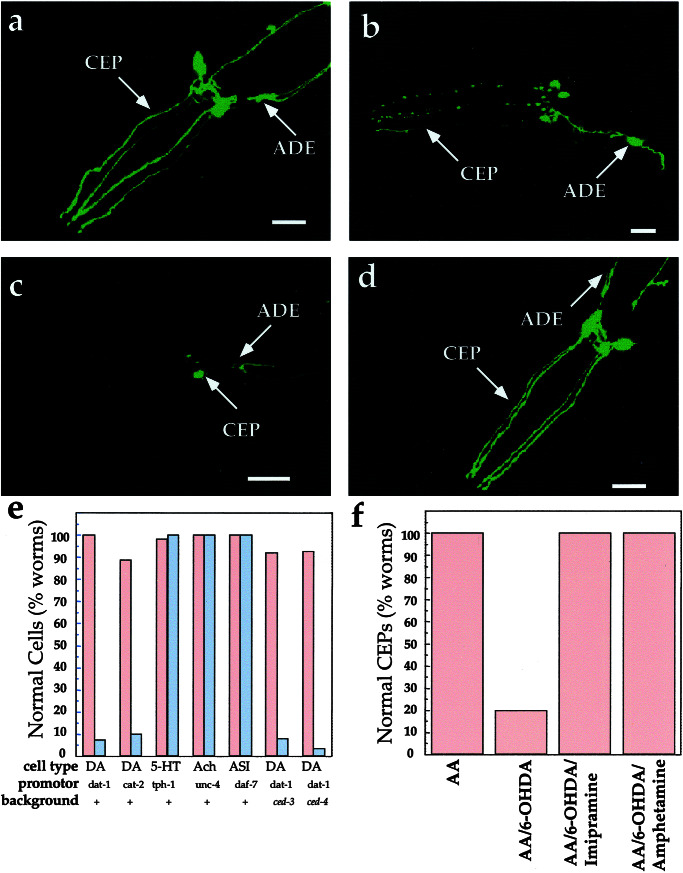
Transfection of DAT-1 cDNA into mammalian cells confers Na+- and Cl−-dependent dopamine transport activity (16). Previous studies demonstrated that dopamine transport in these cells is blocked by nanomolar concentrations of imipramine and nisoxetine (16). In addition, we found that 6-OHDA is a potent inhibitor of [3H]dopamine (50 nM, 5 min, 37°C) transport in vitro, with an IC50 of 341 ± 127 nM (n = 5) in transfected COS cells. To determine whether C. elegans dopamine neurons are sensitive to 6-OHDA in vivo, we exposed nematodes in liquid suspension to a 10 mM 6-OHDA solution for 1 h and then transferred animals to plates lacking toxin. Within 2 h of exposure to the neurotoxin, blebs could be seen along CEP and ADE processes and many of their somas become rounded. By 72 h, we often detected a complete loss of GFP expression in many of the dopamine neurons, with occasional retention of GFP expression in cell bodies (Fig. 2 a–c). Qualitatively similar findings were observed in nonintegrated transgenic lines (data not shown). The different classes of dopamine neurons in the hermaphrodite appeared differentially sensitive to the toxin (CEP>ADE≫PDE) with earliest and most readily detected morphological changes occurring in the CEP processes (data not shown). The loss of GFP signal is unlikely to result from transcriptional repression of the Pdat-1∷GFP transgene, because we achieved quantitatively similar 6-OHDA effects with the Pcat-2∷GFP line (Fig. 2e). Moreover, the 6-OHDA effects appear specific for the dopamine neurons as no morphological changes were evident in nondopaminergic-ergic cells, including serotonergic (5-hydroxytryptamine), cholinergic (acetylcholine), and chemosensory (ASI) neurons visualized with the reporters tph-1∷GFP (37), unc-4∷GFP (38), and daf-7∷GFP, respectively (23) (Fig. 2e).
Figure 2.
Impact of 6-OHDA on dopamine and other neurons. (a) Control worms exposed to 2 mM AA. Morphological changes also were not evident with 10 mM AA. (b) Worms exposed to 50 mM 6-OHDA/10 mM AA; image chosen emphasizes the blebbing along CEP dendrites observed in animals with incomplete penetrance of the 6-OHDA effect. Similar patterns were observed at shorter time points. (c) Worms exposed to 10 mM 6-OHDA/2 mM AA. (d) Worms coexposed to 10 mM 6-OHDA/2 mM AA and 1 mM imipramine. (e) Quantitation of 6-OHDA sensitivity. Animals (≥50 worms/condition) were exposed to either 10 mM AA (red bar) or 50 mM 6-OHDA/10 mM AA (gray bar), and the GFP expression pattern in the respective cells was examined (see Materials and Methods). (f) Quantitation of DAT-1 inhibitor suppression of dopamine neuron sensitivity to 6-OHDA; animals (≥50 worms/condition) were exposed to 2 mM AA, 10 mM 6-OHDA/2 mM AA, and 10 mM 6-OHDA/2 mM AA and either 10 mM d-amphetamine or 1 mM imipramine. For all images, worms are expressing Pdat-1∷GFP. All animals were exposed to 6-OHDA as described in Materials and Methods and examined 3 days postexposure. Anterior is to the left. (All scale bars = 25 μm.) DA, dopamine; Ach, cholinergic neurons (SABV); 5-HT, serotonergic neurons (NSM); ASI, sensory neuron; +, wild type.
Sensitivity of mammalian dopamine neurons in vivo to 6-OHDA is thought to require sequestration of toxin in dopamine terminals and axons by means of presynaptic DATs because toxicity can be prevented by coadministration of DAT antagonists or antisense suppression of DAT expression (39, 40). To determine whether pharmacological suppression of 6-OHDA actions could be observed in C. elegans, we coincubated worms with 6-OHDA or DAT-1 inhibitors (16). Coexposure of worms with 6-OHDA and imipramine (Fig. 2 d and f) or nisoxetine (data not shown) dramatically reduced the loss of GFP expression seen with 6-OHDA alone. The sensitivity of the toxin to the dopamine neurons was also blocked by the psychoactive analog d-amphetamine (Fig. 2f).
Cellular and Ultrastructural Evidence for Dopamine Neuron Degeneration in Nematodes After 6-OHDA Exposure.
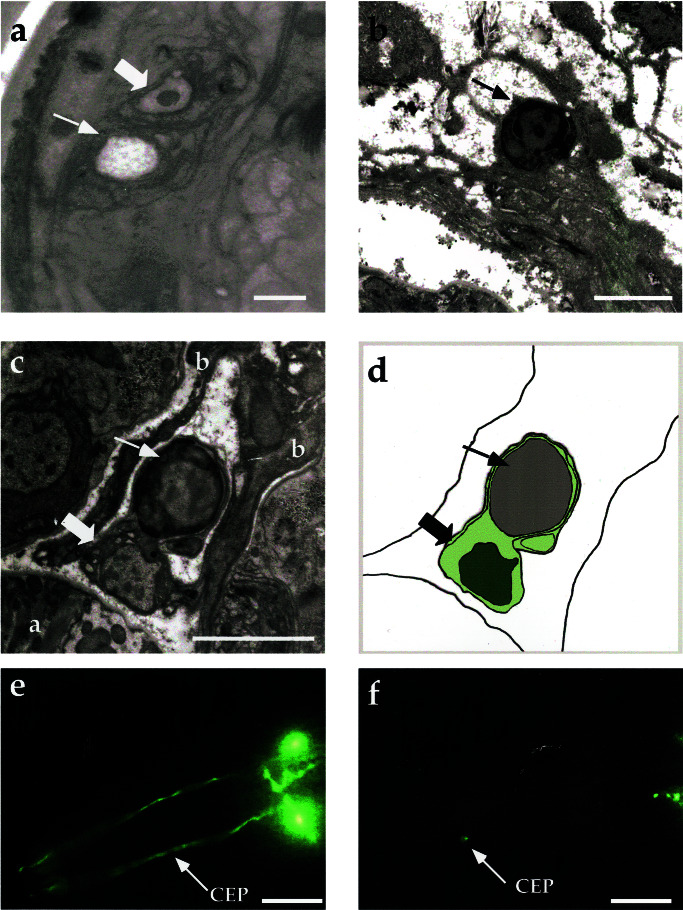
Although the sensitivity of DAT-1-expressing neurons to 6-OHDA appears consistent with cellular insults, we recognized the possibility that nonspecific aggregation or quenching of GFP fluorescence could give a misleading indication of neuronal damage. To better assess morphological aspects of toxin action, we examined the CEP neurons by transmission EM. The positions of all C. elegans cell bodies and processes have been defined by serial section EM reconstruction (32). Wild-type animals were exposed to 6-OHDA as noted above and processed for EM analysis. Serial sections through the CEP dendritic and cell body regions revealed that the majority of the CEP dendritic endings from 6-OHDA-treated worms, but not controls, displayed evident pathological changes. Two of the sensilla were completely absent (2/7), indicating advanced stages of degeneration (Fig. 3a). Many of the dendritic endings that remained appeared shrunken and contained one or two large vacuoles (3/7).
Figure 3.
Ultrastructural and genetic features of cell death in 6-OHDA-treated nematodes. (a) Electron micrograph in the nose of an animal exposed to 50 mM 6-OHDA for 1 h. An open space within the CEP sheath process (thin arrow) is where the CEP dendrite has disappeared. Thick arrow points to the OLQ dendrite, which is normal in appearance. (b) Apoptotic-like, presumptive CEP soma showing condensed chromatin after exposure to 10 mM 6-OHDA for 1 h. The cytoplasm has become more electron dense and the cell is shrunken and rounded, without apparent processes extending from it. (c) Dead, presumptive CEP cell body (thin arrow) engulfed by a GLR cell (thick arrow) (a marks the pharynx, b marks muscle arms). (d) Schematic of c showing engulfment. GLR nucleus is shown in dark green, GLR cytoplasm in light green (all worms prepared for EM were fixed approximately 60 h posttoxin exposure). (e) Representative ced-3(n717) worms exposed to AA (vehicle), and (f) ced-3(n717) exposed to 50 mM 6-OHDA for 1 h. ced-4(n1162) gives identical results (Fig. 2e; data not shown). All 6-OHDA solutions were in 10 mM AA. (e and f) Worms were examined 3 days posttoxin exposure. (Scale bars: a = 0.1 μm; b and c = 0.5 μm; e and f = 25 μm.)
Altogether, 7/12 putative CEP cell bodies showed overt pathology in the three animals examined. The remaining cell bodies (5/12) could not be readily identified, either because they were unaffected or because they had been eliminated. Only one neuron cell body in addition to the presumptive CEPs showed similar pathology in this region (≈1/100 neuron cell bodies examined). Although more extensive analyses at intermediate stages of degeneration are warranted, we found no evidence of membranous whorls, swollen organelles, or swollen cell bodies in injured cells characteristic of necrotic cell death (20). Rather, the damaged cell bodies of presumptive CEPs appeared small, dark, and rounded (Fig. 3b). Their nuclei also appeared dark, with chromatin condensation evident (Fig. 3 b and c), features that are more consistent with an apoptotic pattern of cell death (20, 21). One putative CEP soma was seen in the process of being phagocytosed by a neighboring glial cell (GLR) (Fig. 3 c and d). The GLR could be identified by its prominent cytoplasmic vacuoles and characteristic cell position adjacent to the dorsal right CEP.
Dopamine Neuronal Degeneration Involves CED-3 and CED-4 Independent Pathways.
There is considerable uncertainty as to whether dopamine cell death in PD arises by means of apoptosis, necrosis, neither, or both (41). Direct evidence for cellular features of apoptosis, such as cell shrinkage, nuclear condensation, caspase activation, or nuclear fragmentation may be difficult to achieve in postmortem PD samples because of the relatively brief period of cell degeneration as compared with the protracted course of the disease. Moreover, the existence of multiple members of the ICE family of cysteine proteases (or caspases) involved in programmed cell death in mammals further compounds the difficulty of elucidating molecular pathways. In the nematode, however, programmed cell death is an apoptotic process and is thought to depend on the actions of a single caspase, CED-3, that is closely related to caspase-9 (21). In the nematode, CED-3 is activated by CED-4 (the mammalian APAF-1 homologue) to initiate apoptosis. To determine whether CED-3 and/or CED-4 are required for neurotoxin sensitivity, we crossed our transcriptional Pdat-1∷GFP reporter strain into ced-3 and ced-4 mutant backgrounds to permit visualization of dopamine neurons and treated these animals with 6-OHDA. As with our wild-type reporter strain, dopamine neurons in these mutant lines displayed extensive neuritic blebbing and loss of GFP after toxin treatment; we could detect no differences in the extent of degeneration versus animals with wild-type ced-3 and ced-4 genes (Figs. 2e and 3 e and f). Thus, the programmed cell death pathway defined by CED-3 and CED-4 is not required for 6-OHDA-mediated dopamine neuron degeneration.
Genetic Suppression of 6-OHDA Sensitivity.
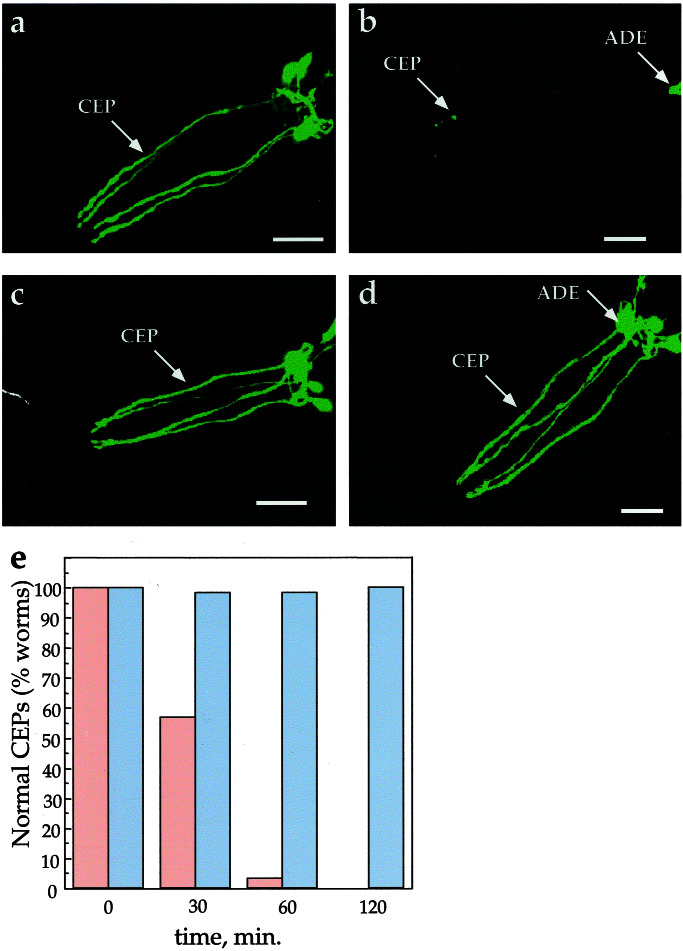
Documentation of 6-OHDA-induced dopamine neuron degeneration in C. elegans raises the possibility of identifying genes that contribute to, or protect against, toxin action. To validate this concept, we tested whether toxin action could be prevented in strains harboring a deleted DAT-1 gene. The mutant, dat-1(ok157), contains a 1.8-kb disruption from exon 4 through exon 12 (J. Duerr and J. Rand, personal communication) and likely results in a null mutation because the majority of the protein (predicted transmembrane domains 4–12), including sites thought crucial for dopamine transport function (42), is deleted. A line homozygous for dat-1(ok157) expressing our transcriptional reporter (Pdat-1∷GFP) was examined for 6-OHDA sensitivity by using confocal fluorescence microscopy as well as EM analysis. As shown in Fig 4, the loss of DAT does not appear to affect the gross morphology of the dopamine neurons nor extinguish the transcription of the dat-1 promoter. However, loss of dat-1 function renders the dopamine neurons insensitive to 6-OHDA (Fig. 4 a–d). GFP fluorescence was not reduced, nor was there any apparent dopamine-neuronal morphological changes even at longer exposure times and at higher toxin concentrations. After a 2-h 6-OHDA exposure, 100% of the CEP neurons are indistinguishable from those found in the nonexposed control worms (Fig. 4e). At the EM level, all dat-1 dendritic endings examined after the 6-OHDA treatment were present. In three worms we examined in detail, all of the dendritic endings were intact (12/12). Two of the endings show one or two very small vacuoles (<50 nm), but the dendrites appear to be otherwise unaffected. Of the three dat-1 worms we examined in detail near the nerve ring, no cells showed abnormal morphology. To verify that loss of DAT accounts for the insensitivity to 6-OHDA in the dat-1 mutant line, we introduced the translational fusion Pdat-1∷DAT-1∷GFP into the dat-1 mutant and found a recovery of 6-OHDA sensitivity (data not shown). Along with our previous demonstration of suppression of 6-OHDA sensitivity through pharmacological DAT-1 blockade, these genetic studies support the hypothesis that 6-OHDA must be transported across the plasma membrane of dopamine neurons to effect degeneration.
Figure 4.
Genetic suppression of 6-OHDA sensitivity of DA neurons. (a) Pdat-1∷GFP animals exposed to 10 mM AA. (b) Pdat-1∷GFP worms exposed to 50 mM 6-OHDA/10 mM AA. (c) Pdat-1∷GFP, dat-1(ok157) worms exposed to 10 mM AA. (d) Pdat-1∷GFP, dat-1(ok157) worms exposed to 50 mM 6-OHDA/10 mM AA (all animals in a–d were exposed for 2 h and examined 3 days later); (e) effect of 50 mM 6-OHDA on the CEP neurons in wild-type and dat-1 worms as a function of time. After a 2-h 6-OHDA exposure (≥50 animals per exposure condition), 100% of the worms in the wild-type background had significant degeneration in their CEPs (red bars; see Materials and Methods for scoring), whereas 100% of the dat-1 worms CEPs appear unaffected by exposure to the toxin (blue bars). (Scale bars = 25 μm.)
Discussion
Visualization of Dopamine Neurons in Living Organisms.
Dopamine neurons can be identified in rodent and primate brain sections by using histofluorescence and immunocytochemical techniques and in vivo by using positron emission tomography (43, 44). For experimental evaluation of dopamine neuron viability in vivo, genomic integration of modified dopamine neuron-specific promoter elements offers the possibility for direct visualization of dopamine neurons as well as the cell-specific introduction of modulatory transgenes. However, elements providing cell-specific expression of DATs in mammalian neurons have not been identified and are likely to be spread over a large span of genomic DNA. Moreover, dopamine neurons and their projections are located deep in the ventral midbrain and basal ganglia of the central nervous system, thereby restricting visualization in intact, adult animals. In contrast to these limitations in mammals, only ≈0.7 kb of 5′ flanking DAT-1 genomic sequence is required to target reporter genes to all eight dopamine neurons within the C. elegans hermaphrodite. As a transparent organism, C. elegans affords the possibility of direct assessment of neuronal integrity by using GFP transgenes in concert with standard fluorescence optics; we have taken this approach to visualize dopamine neurons in the living nematode.
DAT-1 Expression Is Required for 6-OHDA Toxicity.
Previously, we cloned C. elegans DAT-1 cDNA and characterized its substrate specificity and antagonist sensitivity in transfected cells (16). Here we show that 6-OHDA is a potent DAT-1 inhibitor (IC50 = 341 nM), that 6-OHDA affects the viability of C. elegans dopamine neurons, and that loss of DAT-1 function in nematodes, either through pharmacologic blockade (e.g., imipramine, nisoxetine, amphetamine) or genetic disruption abolishes 6-OHDA sensitivity. Preliminary studies also indicate that driving DAT expression in nondopamine neurons may confer sensitivity to 6-OHDA, suggesting opportunities for selective chemical ablation with this toxin (data not shown).
In mammals, DATs are highly expressed by dopamine neurons in the substantia nigra, which project extensively to the caudate and putamen, pathways that are damaged in PD. Within the nigrostriatal projection, DATs are primarily localized perisynaptically along the axonal membrane (43, 45). Interestingly, we detect prominent blebbing of the CEP dendrites after 6-OHDA treatments. These data are consistent with DAT-1 expression in the CEP dendrites (R.N., unpublished findings; J. Duerr and J. Rand, personal communication). In this regard, EM studies of mammalian dopamine neurons also provide evidence of dendritic expression (43). Alternatively, 6-OHDA may diffuse or be transported into the CEP or ADE dendrites to effect toxicity or the dendrites may be damaged through lack of metabolic support from the cell body.
Eventually, extensive damage is seen in all dopamine neuron cell bodies and processes with the notable exception of the PDE neurons. The resistance of PDEs to 6-OHDA may be caused by the inability of the toxin to reach the neuron (i.e., the neurons may be shielded physically or enzymatically within the body cavity) or the PDEs may produce reduced amounts of DAT on the cell surface. Lower amounts of DAT mRNA and protein are expressed by mesolimbic and tuberoinfundibular dopamine neurons, nuclei that also are less affected in PD (46). Alternatively, the PDE neurons may harbor protective genes whose identity could be revealed through genetic screens.
6-OHDA Induced Cell Death May Involve a Novel Apoptotic Pathway.
Ultrastructural analysis of the CEP dendritic endings and cell bodies after exposure to 6-OHDA reveals signs reminiscent of apoptosis. The endings are either absent or vacuolated without any apparent swelling or collateral damage in neighboring cells. The damaged cell bodies are dark and reduced in size, and their chromatin are condensed. Evidence of necrotic cell death was not found; none of the putative dopamine cell bodies examined displayed swelling, and no electron dense whorls were observed within the cytoplasm. These results are consistent with several studies in which the morphological and biochemical correlates suggest apoptosis after treatment with 6-OHDA (47–49). Remarkably, neither the caspase CED-3 nor its activator CED-4, proteins thought to support apoptosis in nematodes, appear to play a role in 6-OHDA-induced dopamine neuron degeneration. The C. elegans genome contains three additional caspase-related genes, which encode several different transcripts (50). To date however, the expression pattern and functions of these proteins in vivo are unknown; they do not appear to participate in any known programmed cell death pathway. It is tempting to speculate that one or more of the undefined caspases may be activated in adult neurons after a toxin-induced insult (e.g., 6-OHDA, or increase in dopamine or reactive oxygen species production). Alternatively, a caspase-independent apoptotic mechanism of cell death may exist. In this regard, the C. elegans genome encodes a gene (GenBank accession no. U50301) highly similar to the mammalian apoptosis inducing factor (AIF), which has been shown to be required for programmed cell death (51). Intriguingly, AIF contains a mitochondrial localization motif and a domain highly homologous to oxidoreductases. Considering that 6-OHDA can induce dopamine neuronal reactive oxygen species production by inhibiting mitochondrial electron transport, this cell death effector could play a significant role in dopamine neurodegeneration.
In summary, we have developed a genetic system to explore mechanisms and therapies of dopamine neuron degeneration. Protection against 6-OHDA effects in the dat-1 line provides essential proof of concept that genes required for toxin-induced neural degeneration can be identified. The mutagenesis of our Pdat-1∷GFP reporter line to screen for intact dopamine neurons after exposure to 6-OHDA will likely allow us to identify molecules involved in DAT regulation and toxin-mediated cell death and is suitable for the study of human genes known to cause rare familiar forms of PD, such as parkin and α-synuclein (17, 52, 53). Finally, our documentation of pharmacologic protection of 6-OHDA-induced dopamine neuron injury raises the prospect that the system will allow for a facile, high-throughput screening approach to identify novel pharmaceutical leads for the treatment of PD.
Supplementary Material
Acknowledgments
We gratefully acknowledge the gift of dat-1(ok157) (RM2704) from J. Rand, the th∷gfp line from R. Lints and S. Emmons, the daf-7∷gfp line from D. Riddle, the tph-1∷gfp line from J. Sze and G. Ruvkun, and GFP expression vectors from A. Fire. We appreciate the technical assistance of M. Schweikhart and Q. Han, imaging consultation with D. Piston and S. Wells, EM sample preparation with G. Stepheney and K. Nguyen, and helpful discussions with D. Greenstein and E. Link. Some strains were provided by the Caenorhabditis Genetics Center, which is supported by the National Institutes of Health National Center for Research Resources. R.D.B. and D.M.M. were supported by National Institutes of Health Grant P01DK58212, R.N. was supported by a PhRMA Foundation Award and a fellowship from the Vanderbilt Training Program in Cellular and Molecular Neuroscience (MH19732–07), and D.H. was supported by National Institutes of Health Grant RR12596. We also acknowledge the generous support of the Vanderbilt University Medical Center Cell Imaging Shared Resource (CA68485 and DK20593).
Abbreviations
- PD
Parkinson's disease
- DAT
dopamine transporter
- GFP
green fluorescent protein
- CEP
cephalic sensilla
- 6-OHDA
6-hydroxydopamine
- ADE
anterior deirids
- PDE
posterior deirids
- EM
electron microscopy
- AA
ascorbic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Yahr M D, Bergman K, editors. Adv Neurol. 1986;45:1–616. [Google Scholar]
- 2.Polymeropoulos M H, Lavedan C, Leroy E, Ide S E, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Nature (London) 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.Jenner P. Movement Disorders. 1998;13:24–34. [PubMed] [Google Scholar]
- 5.McLaughlin B. In: Molecular Mechanisms of Neurodegenerative Disease. Chesselet M, editor. Totowa, NJ: Humana; 2001. pp. 195–231. [Google Scholar]
- 6.Conway K A, Rochet J C, Bieganski R M, Lansbury P T., Jr Science. 2001;294:1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 7.Reading R J, Dunnett S B. In: Toxin-Induced Models of Neurological Disorders. Woodruff M L, Nonnemann A J, editors. New York: Plenum; 1994. pp. 89–119. [Google Scholar]
- 8.Betarbet R, Sherer T B, MacKenzie G, Garcia-Osuna M, Panov A V, Greenamyre J T. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 9.Sachs C, Jonsson G. Biochem Pharmacol. 1975;24:1–8. doi: 10.1016/0006-2952(75)90304-4. [DOI] [PubMed] [Google Scholar]
- 10.Javitch J A, D'Amato R J, Strittmatter S M, Snyder S H. Proc Natl Acad Sci USA. 1985;82:2173–2177. doi: 10.1073/pnas.82.7.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glinka Y, Gassen M, Youdim M B H. J Neural Transm. 1997;50:55–66. doi: 10.1007/978-3-7091-6842-4_7. [DOI] [PubMed] [Google Scholar]
- 12.Curtius H C, Wolfensberger M, Steinmann B, Redweik U, Siegfried J. J Chromatogr. 1974;99:529–540. doi: 10.1016/s0021-9673(00)90882-3. [DOI] [PubMed] [Google Scholar]
- 13.Andrew R, Watson D G, Best S A, Midgley J M, Wenlong H, Petty R K. Neurochem Res. 1993;18:1175–1177. doi: 10.1007/BF00978370. [DOI] [PubMed] [Google Scholar]
- 14.Jellinger K, Linert L, Kienzl E, Herlinger E, Youdim M B. J Neural Transm. 1995;Suppl., 46:297–314. [PubMed] [Google Scholar]
- 15.Sulston J, Dew M, Brenner S. J Comp Neurol. 1975;163:215–226. doi: 10.1002/cne.901630207. [DOI] [PubMed] [Google Scholar]
- 16.Jayanthi L D, Apparsundaram S, Malone M D, Ward E, Miller D M, Eppler M, Blakely R D. Mol Pharmacol. 1998;54:601–609. [PubMed] [Google Scholar]
- 17.Nass R, Miller D M, Blakely R D. Parkinsonism Related Disorders. 2001;7:185–191. doi: 10.1016/s1353-8020(00)00056-0. [DOI] [PubMed] [Google Scholar]
- 18.Duerr J S, Frisby D L, Gaskin J, Duke A, Asermely K, Huddleston D, Eiden L E, Rand J B. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rand J B, Nonet M L. In: in C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 611–643. [Google Scholar]
- 20.Hall D H, Gu G, Garcia-Anoveros J, Gong L, Chalfie M, Driscoll M. J Neurosci. 1997;17:1033–1045. doi: 10.1523/JNEUROSCI.17-03-01033.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengartner M O. In: in C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Press J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 383–415. [Google Scholar]
- 22.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren P, Lim C S, Johnsen R, Albert P S, Pilgrim D, Riddle D L. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 24.Yuan J, Horvitz H R. Development (Cambridge, UK) 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Shaham S, Ledoux S, Ellis H M, Horvitz H R. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 26.Shaham S, Reddien P W, Davies B, Horvitz H R. Genetics. 1999;153:1655–1671. doi: 10.1093/genetics/153.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. Nature (London) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 28.Lynch A S, Briggs D, Hope I A. Nat Genet. 1995;11:309–313. doi: 10.1038/ng1195-309. [DOI] [PubMed] [Google Scholar]
- 29.Kramer J M, French R P, Park E C, Johnson J J. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mello C, Fire A. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 31.Lewis J A, Fleming J T. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 32.White J G, Southgate E, Thompson J N, Brenner S. Philos Trans R Soc London B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 33.Schafer W R, Kenyon C J. Nature (London) 1995;375:73–78. doi: 10.1038/375073a0. [DOI] [PubMed] [Google Scholar]
- 34.Sawin E R, Ranganathan R, Horvitz H R. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 35.Ranganathan R, Sawin E R, Trent C, Horvitz H R. J Neurosci. 2001;21:5871–5884. doi: 10.1523/JNEUROSCI.21-16-05871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sulston J E, Horvitz H R. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 37.Sze J Y, Victor M, Loer C, Shi Y, Ruvkun G. Nature (London) 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- 38.Winnier A R, Meir J Y, Ross J M, Tavernarakis N, Driscoll M, Ishihara T, Katsura I, Miller D M., 3rd Genes Dev. 1999;13:2774–2786. doi: 10.1101/gad.13.21.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waddington J L. Pharmacol Biochem Behav. 1980;13:915–917. doi: 10.1016/0091-3057(80)90229-4. [DOI] [PubMed] [Google Scholar]
- 40.Van Kampen J M, McGeer E G, Stoessl A J. Synapse. 2000;37:171–178. doi: 10.1002/1098-2396(20000901)37:3<171::AID-SYN1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 41.Jellinger K A. J Neural Transm. 2000;107:1–29. doi: 10.1007/s007020050001. [DOI] [PubMed] [Google Scholar]
- 42.Giros B, Wang Y M, Suter S, McLeskey S B, Pifl C, Caron M G. J Biol Chem. 1994;269:15985–15988. [PubMed] [Google Scholar]
- 43.Nirenberg M J, Vaughan R A, Uhl G R, Kuhar M J, Pickel V M. J Neurosci. 1996;16:436–447. doi: 10.1523/JNEUROSCI.16-02-00436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hershey T, Moerlein S M, Perlmutter J S. In: Molecular Mechanisms of Neurodegenerative Diseases. Chesselet M, editor. Totowa, NJ: Humana; 2001. pp. 177–193. [Google Scholar]
- 45.Pickel V M, Nirenberg M J, Milner T A. J Neurocytol. 1996;25:843–856. doi: 10.1007/BF02284846. [DOI] [PubMed] [Google Scholar]
- 46.Uhl G R. Ann Neurol. 1998;43:555–560. doi: 10.1002/ana.410430503. [DOI] [PubMed] [Google Scholar]
- 47.Choi W S, Yoon S Y, Oh T H, Choi E J, O'Malley K L, Oh Y J. J Neurosci Res. 1999;57:86–94. doi: 10.1002/(SICI)1097-4547(19990701)57:1<86::AID-JNR9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 48.Dodel R C, Du Y, Bales K R, Ling Z, Carvey P M, Paul S M. Brain Res Mol Brain Res. 1999;64:141–148. doi: 10.1016/s0169-328x(98)00318-0. [DOI] [PubMed] [Google Scholar]
- 49.Lotharius J, Dugan L L, O'Malley K L. J Neurosci. 1999;19:1284–1293. doi: 10.1523/JNEUROSCI.19-04-01284.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaham S. J Biol Chem. 1998;273:35109–35117. doi: 10.1074/jbc.273.52.35109. [DOI] [PubMed] [Google Scholar]
- 51.Lorenzo H K, Susin S A, Penninger J, Kroemer G. Cell Death Differ. 1999;6:516–524. doi: 10.1038/sj.cdd.4400527. [DOI] [PubMed] [Google Scholar]
- 52.Feany M B, Bender W W. Nature (London) 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 53.Lee F J, Liu F, Pristupa Z B, Niznik H B. FASEB J. 2001;15:916–926. doi: 10.1096/fj.00-0334com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.