Abstract
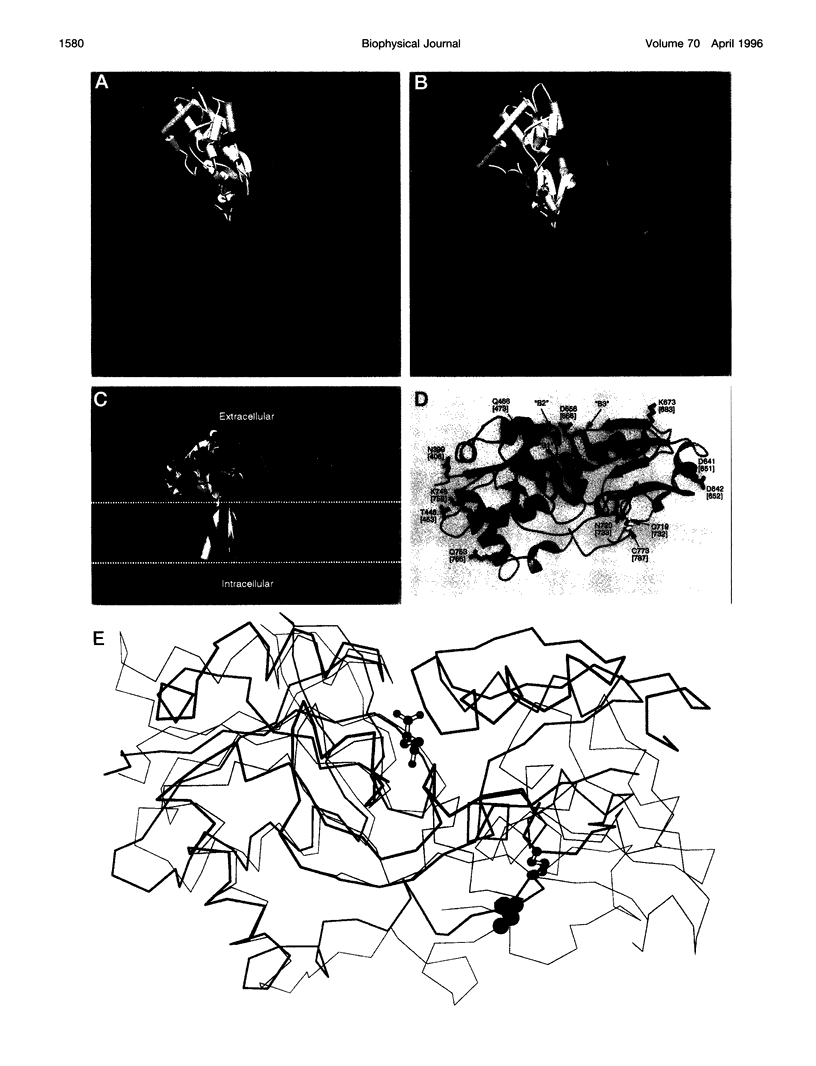
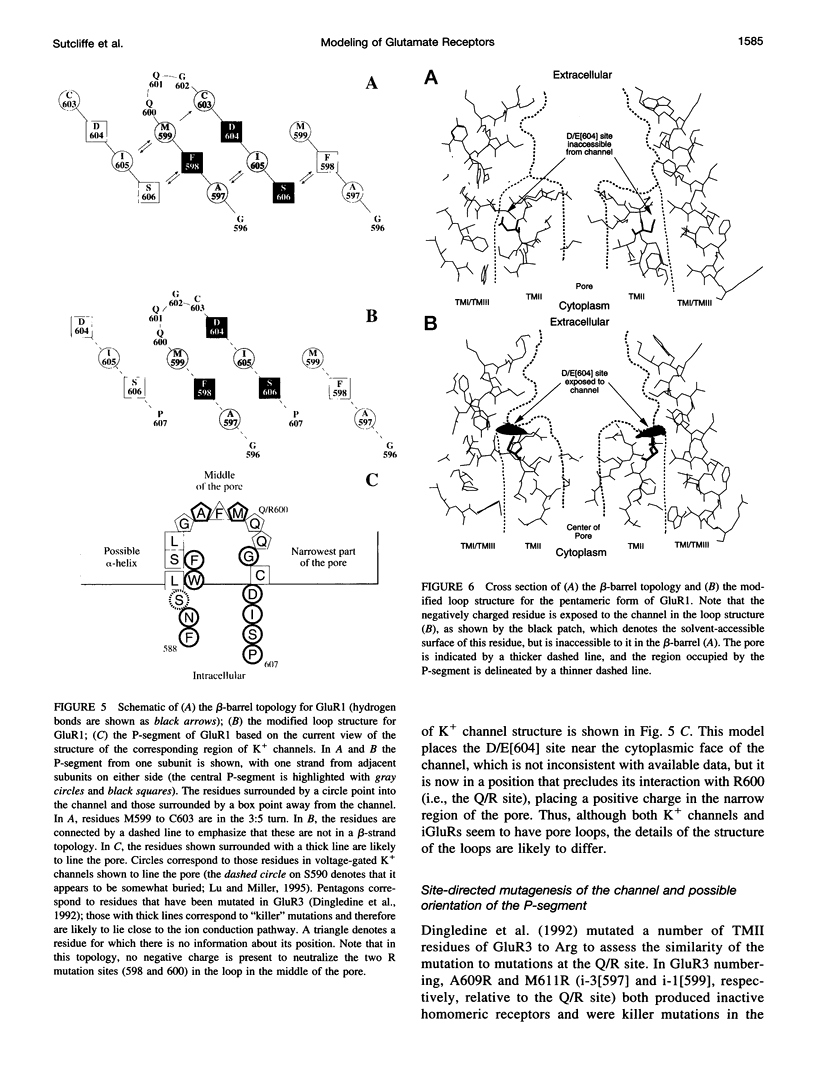
Structural models have been produced for three types of non-NMDA inotropic glutamate receptors: an AMPA receptor, GluR1, a kainate receptor, GluR6; and a low-molecular-weight kainate receptor from goldfish, GFKAR alpha. Modeling was restricted to the domains of the proteins that bind the neurotransmitter glutamate and that form the ion channel. Model building combined homology modeling, distance geometry, molecular mechanics, interactive modeling, and known constraints. The models indicate new potential interactions in the extracellular domain between protein and agonists, and suggest that the transition from the "closed" to the "open" state involves the movement of a conserved positive residue away from, and two conserved negative residues into, the extracellular entrance to the pore upon binding. As a first approximation, the ion channel domain was modeled with a structure comprising a central antiparallel beta-barrel that partially crosses the membrane, and against which alpha-helices from each subunit are packed; a third alpha-helix packs against these two helices in each subunit. Much, but not all, of the available data were consistent with this structure. Modifying the beta-barrel to a loop-like topology produced a model consistent with available data.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barton G. J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993 Jan;6(1):37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Bennett J. A., Dingledine R. Topology profile for a glutamate receptor: three transmembrane domains and a channel-lining reentrant membrane loop. Neuron. 1995 Feb;14(2):373–384. doi: 10.1016/0896-6273(95)90293-7. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blackstone C. D., Moss S. J., Martin L. J., Levey A. I., Price D. L., Huganir R. L. Biochemical characterization and localization of a non-N-methyl-D-aspartate glutamate receptor in rat brain. J Neurochem. 1992 Mar;58(3):1118–1126. doi: 10.1111/j.1471-4159.1992.tb09370.x. [DOI] [PubMed] [Google Scholar]
- Blundell T., Carney D., Gardner S., Hayes F., Howlin B., Hubbard T., Overington J., Singh D. A., Sibanda B. L., Sutcliffe M. 18th Sir Hans Krebs lecture. Knowledge-based protein modelling and design. Eur J Biochem. 1988 Mar 15;172(3):513–520. doi: 10.1111/j.1432-1033.1988.tb13917.x. [DOI] [PubMed] [Google Scholar]
- Bowie D., Mayer M. L. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995 Aug;15(2):453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Brose N., Gasic G. P., Vetter D. E., Sullivan J. M., Heinemann S. F. Protein chemical characterization and immunocytochemical localization of the NMDA receptor subunit NMDA R1. J Biol Chem. 1993 Oct 25;268(30):22663–22671. [PubMed] [Google Scholar]
- Burnashev N., Monyer H., Seeburg P. H., Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992 Jan;8(1):189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Burnashev N., Schoepfer R., Monyer H., Ruppersberg J. P., Günther W., Seeburg P. H., Sakmann B. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science. 1992 Sep 4;257(5075):1415–1419. doi: 10.1126/science.1382314. [DOI] [PubMed] [Google Scholar]
- Changeux J. P., Galzi J. L., Devillers-Thiéry A., Bertrand D. The functional architecture of the acetylcholine nicotinic receptor explored by affinity labelling and site-directed mutagenesis. Q Rev Biophys. 1992 Nov;25(4):395–432. doi: 10.1017/s0033583500004352. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Epp O., Sinning I., Michel H. Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis. J Mol Biol. 1995 Feb 24;246(3):429–457. doi: 10.1006/jmbi.1994.0097. [DOI] [PubMed] [Google Scholar]
- Dingledine R., Hume R. I., Heinemann S. F. Structural determinants of barium permeation and rectification in non-NMDA glutamate receptor channels. J Neurosci. 1992 Oct;12(10):4080–4087. doi: 10.1523/JNEUROSCI.12-10-04080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durell S. R., Guy H. R. Atomic scale structure and functional models of voltage-gated potassium channels. Biophys J. 1992 Apr;62(1):238–250. doi: 10.1016/S0006-3495(92)81809-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ealick S. E., Babu Y. S., Bugg C. E., Erion M. D., Guida W. C., Montgomery J. A., Secrist J. A., 3rd Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11540–11544. doi: 10.1073/pnas.88.24.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J., Bettler B., Hermans-Borgmeyer I., Heinemann S. Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not AMPA. Nature. 1991 Jun 27;351(6329):745–748. doi: 10.1038/351745a0. [DOI] [PubMed] [Google Scholar]
- Eismann E., Müller F., Heinemann S. H., Kaupp U. B. A single negative charge within the pore region of a cGMP-gated channel controls rectification, Ca2+ blockage, and ionic selectivity. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):1109–1113. doi: 10.1073/pnas.91.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S. V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993 Jun;11(2):134-8, 127-8. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Ferrer-Montiel A. V., Sun W., Montal M. Molecular design of the N-methyl-D-aspartate receptor binding site for phencyclidine and dizolcipine. Proc Natl Acad Sci U S A. 1995 Aug 15;92(17):8021–8025. doi: 10.1073/pnas.92.17.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger J. R., Melcher T., Koh D. S., Sakmann B., Seeburg P. H., Jonas P., Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995 Jul;15(1):193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Goulding E. H., Tibbs G. R., Liu D., Siegelbaum S. A. Role of H5 domain in determining pore diameter and ion permeation through cyclic nucleotide-gated channels. Nature. 1993 Jul 1;364(6432):61–64. doi: 10.1038/364061a0. [DOI] [PubMed] [Google Scholar]
- Gregor P., Mano I., Maoz I., McKeown M., Teichberg V. I. Molecular structure of the chick cerebellar kainate-binding subunit of a putative glutamate receptor. Nature. 1989 Dec 7;342(6250):689–692. doi: 10.1038/342689a0. [DOI] [PubMed] [Google Scholar]
- Heginbotham L., Abramson T., MacKinnon R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 1992 Nov 13;258(5085):1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Henley J. M., Oswald R. E. Solubilization and characterization of kainate receptors from goldfish brain. Biochim Biophys Acta. 1988 Jan 13;937(1):103–111. doi: 10.1016/0005-2736(88)90232-5. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hollmann M., Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Hollmann M., Maron C., Heinemann S. N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994 Dec;13(6):1331–1343. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
- Hollmann M., O'Shea-Greenfield A., Rogers S. W., Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989 Dec 7;342(6250):643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- Hume R. I., Dingledine R., Heinemann S. F. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991 Aug 30;253(5023):1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- Imoto K., Busch C., Sakmann B., Mishina M., Konno T., Nakai J., Bujo H., Mori Y., Fukuda K., Numa S. Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature. 1988 Oct 13;335(6191):645–648. doi: 10.1038/335645a0. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994 Mar 15;33(10):3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Keinänen K., Wisden W., Sommer B., Werner P., Herb A., Verdoorn T. A., Sakmann B., Seeburg P. H. A family of AMPA-selective glutamate receptors. Science. 1990 Aug 3;249(4968):556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Kuryatov A., Laube B., Betz H., Kuhse J. Mutational analysis of the glycine-binding site of the NMDA receptor: structural similarity with bacterial amino acid-binding proteins. Neuron. 1994 Jun;12(6):1291–1300. doi: 10.1016/0896-6273(94)90445-6. [DOI] [PubMed] [Google Scholar]
- Köhler M., Burnashev N., Sakmann B., Seeburg P. H. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993 Mar;10(3):491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Kürz L. L., Zühlke R. D., Zhang H. J., Joho R. H. Side-chain accessibilities in the pore of a K+ channel probed by sulfhydryl-specific reagents after cysteine-scanning mutagenesis. Biophys J. 1995 Mar;68(3):900–905. doi: 10.1016/S0006-3495(95)80266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Owens N., Verdoorn T. A. Functional effects of mutations in the putative agonist binding region of recombinant alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors. Mol Pharmacol. 1995 Jan;47(1):148–154. [PubMed] [Google Scholar]
- Liman E. R., Tytgat J., Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992 Nov;9(5):861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- Lü Q., Miller C. Silver as a probe of pore-forming residues in a potassium channel. Science. 1995 Apr 14;268(5208):304–307. doi: 10.1126/science.7716526. [DOI] [PubMed] [Google Scholar]
- MacKinnon R. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature. 1991 Mar 21;350(6315):232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- MacKinnon R. Pore loops: an emerging theme in ion channel structure. Neuron. 1995 May;14(5):889–892. doi: 10.1016/0896-6273(95)90327-5. [DOI] [PubMed] [Google Scholar]
- Merritt E. A., Murphy M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994 Nov 1;50(Pt 6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Monaghan D. T., Bridges R. J., Cotman C. W. The excitatory amino acid receptors: their classes, pharmacology, and distinct properties in the function of the central nervous system. Annu Rev Pharmacol Toxicol. 1989;29:365–402. doi: 10.1146/annurev.pa.29.040189.002053. [DOI] [PubMed] [Google Scholar]
- Nakanishi N., Shneider N. A., Axel R. A family of glutamate receptor genes: evidence for the formation of heteromultimeric receptors with distinct channel properties. Neuron. 1990 Nov;5(5):569–581. doi: 10.1016/0896-6273(90)90212-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S., Masu M. Molecular diversity and functions of glutamate receptors. Annu Rev Biophys Biomol Struct. 1994;23:319–348. doi: 10.1146/annurev.bb.23.060194.001535. [DOI] [PubMed] [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992 Oct 23;258(5082):597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- O'Hara P. J., Sheppard P. O., Thøgersen H., Venezia D., Haldeman B. A., McGrane V., Houamed K. M., Thomsen C., Gilbert T. L., Mulvihill E. R. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993 Jul;11(1):41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- Oh B. H., Pandit J., Kang C. H., Nikaido K., Gokcen S., Ames G. F., Kim S. H. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J Biol Chem. 1993 May 25;268(15):11348–11355. [PubMed] [Google Scholar]
- Pascual J. M., Shieh C. C., Kirsch G. E., Brown A. M. K+ pore structure revealed by reporter cysteines at inner and outer surfaces. Neuron. 1995 May;14(5):1055–1063. doi: 10.1016/0896-6273(95)90344-5. [DOI] [PubMed] [Google Scholar]
- Persson B., Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994 Mar 25;237(2):182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Roche K. W., Raymond L. A., Blackstone C., Huganir R. L. Transmembrane topology of the glutamate receptor subunit GluR6. J Biol Chem. 1994 Apr 22;269(16):11679–11682. [PubMed] [Google Scholar]
- Root M. J., MacKinnon R. Identification of an external divalent cation-binding site in the pore of a cGMP-activated channel. Neuron. 1993 Sep;11(3):459–466. doi: 10.1016/0896-6273(93)90150-p. [DOI] [PubMed] [Google Scholar]
- Root M. J., MacKinnon R. Two identical noninteracting sites in an ion channel revealed by proton transfer. Science. 1994 Sep 23;265(5180):1852–1856. doi: 10.1126/science.7522344. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994 May;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Sibanda B. L., Blundell T. L., Thornton J. M. Conformation of beta-hairpins in protein structures. A systematic classification with applications to modelling by homology, electron density fitting and protein engineering. J Mol Biol. 1989 Apr 20;206(4):759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- Sippl M. J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993 Dec;17(4):355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Smart O. S., Goodfellow J. M., Wallace B. A. The pore dimensions of gramicidin A. Biophys J. 1993 Dec;65(6):2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Keinänen K., Verdoorn T. A., Wisden W., Burnashev N., Herb A., Köhler M., Takagi T., Sakmann B., Seeburg P. H. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990 Sep 28;249(4976):1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., Seeburg P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991 Oct 4;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Stanfield P. R., Davies N. W., Shelton P. A., Sutcliffe M. J., Khan I. A., Brammar W. J., Conley E. C. A single aspartate residue is involved in both intrinsic gating and blockage by Mg2+ of the inward rectifier, IRK1. J Physiol. 1994 Jul 1;478(Pt 1):1–6. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Bach Y., Bettler B., Hartley M., Sheppard P. O., O'Hara P. J., Heinemann S. F. Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron. 1994 Dec;13(6):1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- Sullivan J. M., Traynelis S. F., Chen H. S., Escobar W., Heinemann S. F., Lipton S. A. Identification of two cysteine residues that are required for redox modulation of the NMDA subtype of glutamate receptor. Neuron. 1994 Oct;13(4):929–936. doi: 10.1016/0896-6273(94)90258-5. [DOI] [PubMed] [Google Scholar]
- Surles M. C., Richardson J. S., Richardson D. C., Brooks F. P., Jr Sculpting proteins interactively: continual energy minimization embedded in a graphical modeling system. Protein Sci. 1994 Feb;3(2):198–210. doi: 10.1002/pro.5560030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna F. A., Wang L. Y., MacDonald J. F., Hampson D. R. A transmembrane model for an ionotropic glutamate receptor predicted on the basis of the location of asparagine-linked oligosaccharides. J Biol Chem. 1994 May 13;269(19):14159–14164. [PubMed] [Google Scholar]
- Taylor W. R. A flexible method to align large numbers of biological sequences. J Mol Evol. 1988 Dec;28(1-2):161–169. doi: 10.1007/BF02143508. [DOI] [PubMed] [Google Scholar]
- Uchino S., Sakimura K., Nagahari K., Mishina M. Mutations in a putative agonist binding region of the AMPA-selective glutamate receptor channel. FEBS Lett. 1992 Aug 24;308(3):253–257. doi: 10.1016/0014-5793(92)81286-u. [DOI] [PubMed] [Google Scholar]
- Verdoorn T. A., Burnashev N., Monyer H., Seeburg P. H., Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991 Jun 21;252(5013):1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- Villarroel A., Burnashev N., Sakmann B. Dimensions of the narrow portion of a recombinant NMDA receptor channel. Biophys J. 1995 Mar;68(3):866–875. doi: 10.1016/S0006-3495(95)80263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Dechesne C. J., Shimasaki S., King R. G., Kusano K., Buonanno A., Hampson D. R., Banner C., Wenthold R. J., Nakatani Y. Sequence and expression of a frog brain complementary DNA encoding a kainate-binding protein. Nature. 1989 Dec 7;342(6250):684–689. doi: 10.1038/342684a0. [DOI] [PubMed] [Google Scholar]
- Wenthold R. J., Yokotani N., Doi K., Wada K. Immunochemical characterization of the non-NMDA glutamate receptor using subunit-specific antibodies. Evidence for a hetero-oligomeric structure in rat brain. J Biol Chem. 1992 Jan 5;267(1):501–507. [PubMed] [Google Scholar]
- Wisden W., Seeburg P. H. Mammalian ionotropic glutamate receptors. Curr Opin Neurobiol. 1993 Jun;3(3):291–298. doi: 10.1016/0959-4388(93)90120-n. [DOI] [PubMed] [Google Scholar]
- Wo Z. G., Bian Z. C., Oswald R. E. Asn-265 of frog kainate binding protein is a functional glycosylation site: implications for the transmembrane topology of glutamate receptors. FEBS Lett. 1995 Jul 17;368(2):230–234. doi: 10.1016/0014-5793(95)00655-s. [DOI] [PubMed] [Google Scholar]
- Wo Z. G., Oswald R. E. A topological analysis of goldfish kainate receptors predicts three transmembrane segments. J Biol Chem. 1995 Feb 3;270(5):2000–2009. doi: 10.1074/jbc.270.5.2000. [DOI] [PubMed] [Google Scholar]
- Wo Z. G., Oswald R. E. Transmembrane topology of two kainate receptor subunits revealed by N-glycosylation. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7154–7158. doi: 10.1073/pnas.91.15.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wo Z. G., Oswald R. E. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995 Apr;18(4):161–168. doi: 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- Wood M. W., VanDongen H. M., VanDongen A. M. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4882–4886. doi: 10.1073/pnas.92.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhull A. M. Ionic blockage of sodium channels in nerve. J Gen Physiol. 1973 Jun;61(6):687–708. doi: 10.1085/jgp.61.6.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. Y., Chang Y. C. Hydrodynamic and pharmacological characterization of putative alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid/kainate-sensitive L-glutamate receptors solubilized from pig brain. Biochem J. 1994 Jun 1;300(Pt 2):365–371. doi: 10.1042/bj3000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N., Trakhanov S., Quiocho F. A. Refined 1.89-A structure of the histidine-binding protein complexed with histidine and its relationship with many other active transport/chemosensory proteins. Biochemistry. 1994 Apr 26;33(16):4769–4779. doi: 10.1021/bi00182a004. [DOI] [PubMed] [Google Scholar]
- Zarei M. M., Dani J. A. Structural basis for explaining open-channel blockade of the NMDA receptor. J Neurosci. 1995 Feb;15(2):1446–1454. doi: 10.1523/JNEUROSCI.15-02-01446.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]