Abstract
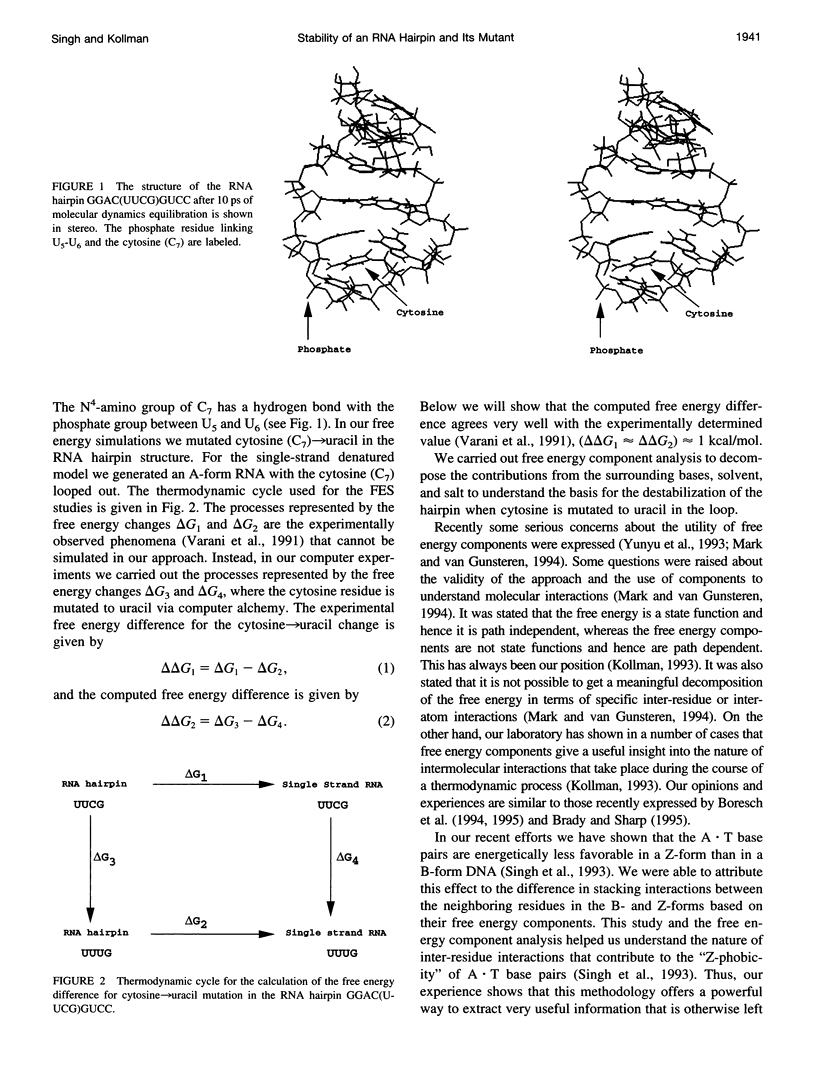
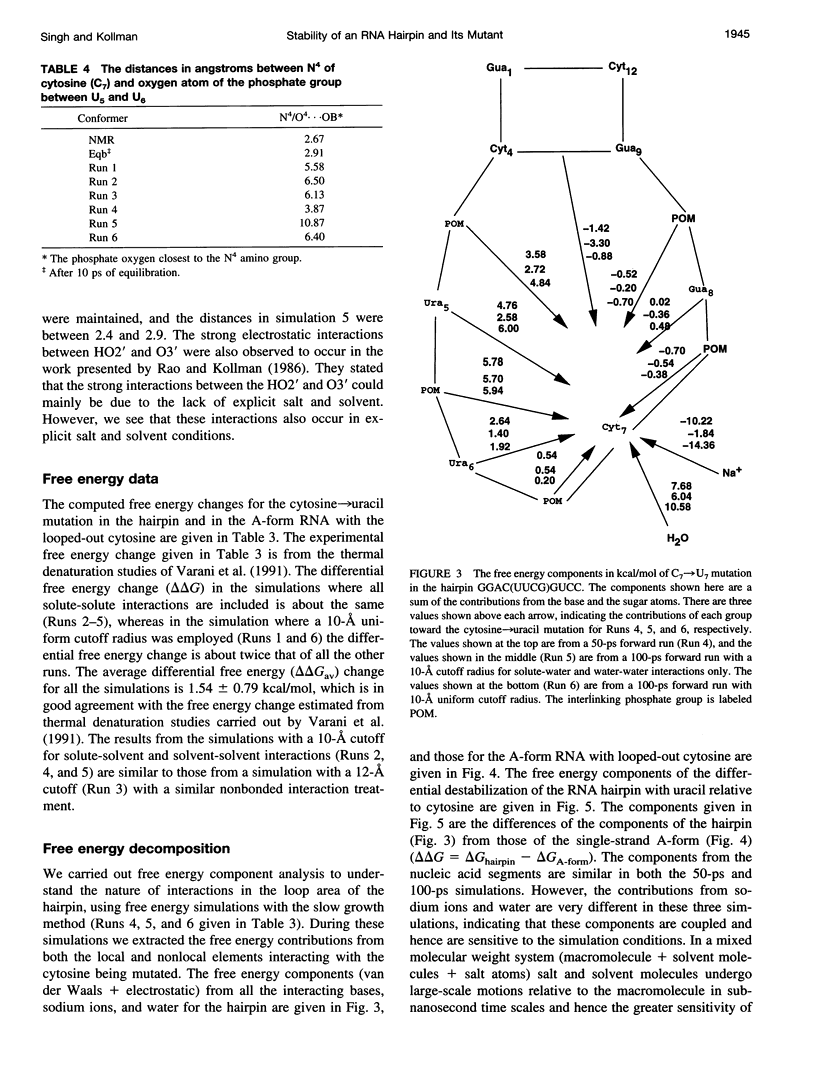
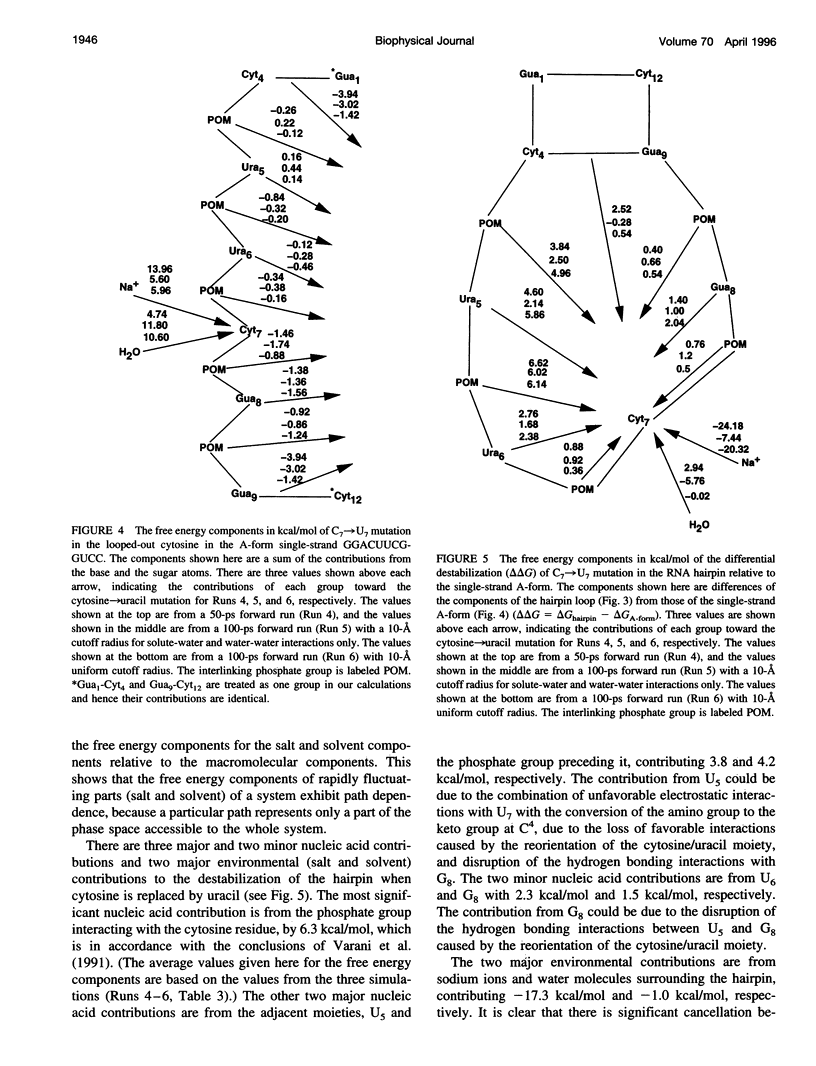
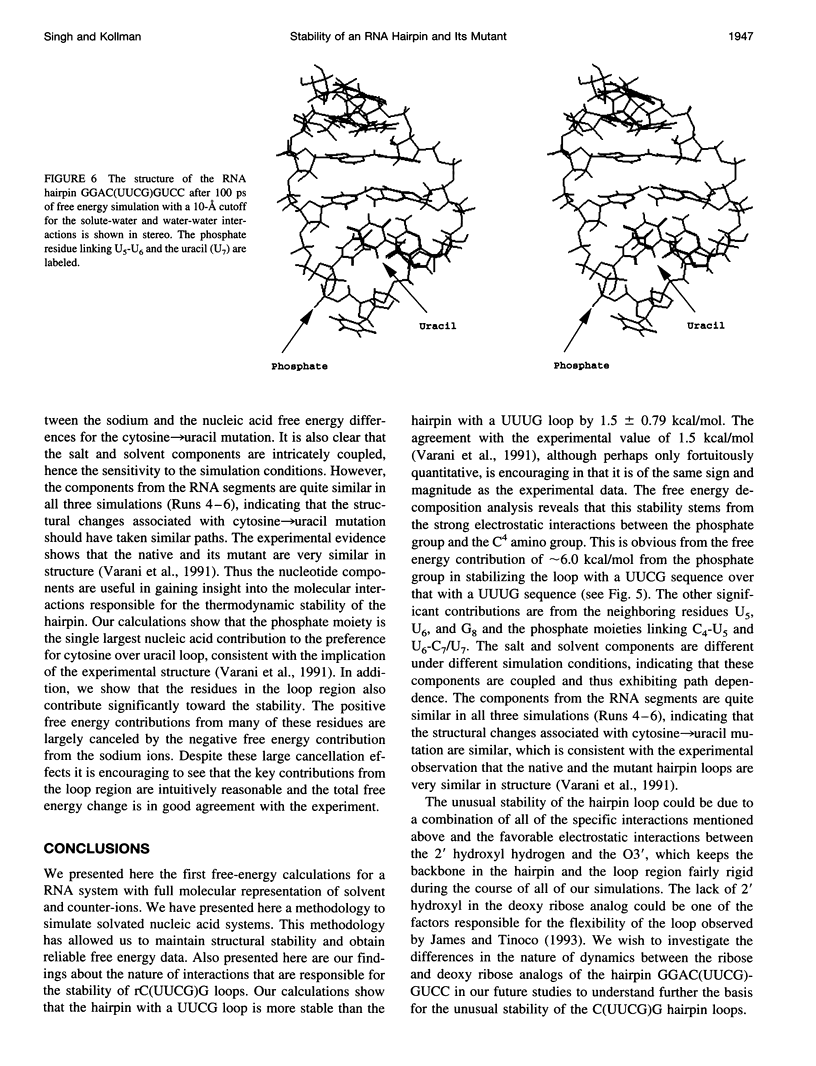
The thermodynamic stability of RNA hairpin loops has been a subject of considerable interest in the recent past (Wimberly et al., 1991). There have been experimental reports indicating that the hairpins with a C(UUCG)G loop sequence are thermodynamically very stable (Wimberly et al., 1991). We used the solution structure of GGAC(UUCG)GUCC (Cheong et al., 1990; Varani et al., 1991) as the starting conformation in our attempt to understand its thermodynamic stability. We carried out molecular dynamics/free energy simulations to understand the basis for the destabilization of the C(UUCG)G loop by mutating cytosine (C7)-->uracil. Because of the limited length of simulation and the presence of kinetic barriers (solvent intervention) to the uracil-->cytosine mutation, all of our computed free energy differences are based on multiple forward simulations. Based on these calculations we find that the cytosine-->uracil mutation in the loop destabilizes it by approximately 1.5kcal/mol relative to that of the reference state, an A-form RNA but with cytosine (C7) looped out. This is the same sign and magnitude as that observed in the thermodynamic studies carried out by Varani et al.(1991). We have carried out free energy component analysis to understand the effect of mutating the cytosine residue to uracil on the thermodynamic stability of the C(UUCG)G hairpin loops. Our calculations show that the most significant contribution to the stability is from the phosphate group linking U5 and U6, which favors the cytosine residue over uracil by about 6.0 kcal/mol. The residues U5, U6, and G8 in the loop region also contribute significantly to the stability. The contributions from the salt and solvent compensate each other, indicating the dynamic nature of interactions of the environment with the nucleic acid system and the coupling between these two components.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W., Dover S. D., Fuller W., Hodgson A. R. Structures of synthetic polynucleotides in the A-RNA and A'-RNA conformations: x-ray diffraction analyses of the molecular conformations of polyadenylic acid--polyuridylic acid and polyinosinic acid--polycytidylic acid. J Mol Biol. 1973 Dec 5;81(2):107–122. doi: 10.1016/0022-2836(73)90183-6. [DOI] [PubMed] [Google Scholar]
- Boresch S., Archontis G., Karplus M. Free energy simulations: the meaning of the individual contributions from a component analysis. Proteins. 1994 Sep;20(1):25–33. doi: 10.1002/prot.340200105. [DOI] [PubMed] [Google Scholar]
- Boresch S., Karplus M. The meaning of component analysis: decomposition of the free energy in terms of specific interactions. J Mol Biol. 1995 Dec 15;254(5):801–807. doi: 10.1006/jmbi.1995.0656. [DOI] [PubMed] [Google Scholar]
- Brady G. P., Sharp K. A. Decomposition of interaction free energies in proteins and other complex systems. J Mol Biol. 1995 Nov 17;254(1):77–85. doi: 10.1006/jmbi.1995.0600. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- James J. K., Tinoco I., Jr The solution structure of a d[C(TTCG)G] DNA hairpin and comparison to the unusually stable RNA analogue. Nucleic Acids Res. 1993 Jul 11;21(14):3287–3293. doi: 10.1093/nar/21.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark A. E., van Gunsteren W. F. Decomposition of the free energy of a system in terms of specific interactions. Implications for theoretical and experimental studies. J Mol Biol. 1994 Jul 8;240(2):167–176. doi: 10.1006/jmbi.1994.1430. [DOI] [PubMed] [Google Scholar]
- Nilsson L., Ahgren-Stålhandske A., Sjögren A. S., Hahne S., Sjöberg B. M. Three-dimensional model and molecular dynamics simulation of the active site of the self-splicing intervening sequence of the bacteriophage T4 nrdB messenger RNA. Biochemistry. 1990 Nov 13;29(45):10317–10322. doi: 10.1021/bi00497a005. [DOI] [PubMed] [Google Scholar]
- Shi Y. Y., Mark A. E., Wang C. X., Huang F., Berendsen H. J., van Gunsteren W. F. Can the stability of protein mutants be predicted by free energy calculations? Protein Eng. 1993 Apr;6(3):289–295. doi: 10.1093/protein/6.3.289. [DOI] [PubMed] [Google Scholar]
- Singh S. B., Pearlman D. A., Kollman P. A. Free energy component analysis: application to the "Z-phobicity" of A-T base pairs. J Biomol Struct Dyn. 1993 Oct;11(2):303–311. doi: 10.1080/07391102.1993.10508728. [DOI] [PubMed] [Google Scholar]
- Singh S. B., Wemmer D. E., Kollman P. A. Relative binding affinities of distamycin and its analog to d(CGCAAGTTGGC).d(GCCAACTTGCG): comparison of simulation results with experiment. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7673–7677. doi: 10.1073/pnas.91.16.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Cheong C., Tinoco I., Jr Structure of an unusually stable RNA hairpin. Biochemistry. 1991 Apr 2;30(13):3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Cech T. R. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986 Jan 31;231(4737):470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]


