Abstract
The light activation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in vivo requires the presence of Rubisco activase, a nuclear-encoded chloroplast protein that consists of two isoforms arising from alternative splicing in most plants. We examined the function of each isoform by characterizing Rubisco activation in transgenic Arabidopsis plants that express only one or both isoforms, as compared with the wild type. In plants expressing only the shorter isoform, Rubisco activity was as high as in the wild type under saturating light, but the activity was not down-regulated at intensities limiting for photosynthesis. In contrast, in plants expressing only the longer isoform, Rubisco activity was down-regulated at limiting light, but the activity was slightly lower and increased much more slowly at saturating light intensities as compared with the wild type. Light regulation of Rubisco similar to that in the wild-type plants was observed in the progeny of a genetic cross of these two transformants in which both isoforms were again present. When the capacity to redox regulate the activity of the larger activase isoform was eliminated by replacement of the critical cysteine residues in the carboxyl-terminal extension unique to this isoform, Rubisco activity in saturating light was similar to the wild type, but the ability of the larger isoform to down-regulate Rubisco activity at limiting light intensities in transgenic plants was almost abolished. These results indicate that the light modulation of Rubisco under limiting light is mainly due to the ability to regulate the activity of Rubisco activase by redox changes in the stroma.
Photosynthetic carbon metabolism is initiated by Rubisco, which combines CO2 with ribulose 1,5-bisphosphate (RuBP) to form two molecules of phosphoglyeric acid. Both photosynthesis and the activity of Rubisco vary with light intensity, which supplies the energy for regeneration of the RuBP substrate. The changes in the activity of Rubisco are caused by “activation” by carbamoylation of a lysine residue and Mg2+-binding in the active site (1). Activity sufficient for high rates of photosynthesis and growth at atmospheric concentrations of CO2 depends on the activity of another protein, Rubisco activase (reviewed in refs. 2 and 3). The activase hydrolyzes ATP to promote the dissociation of inhibitory sugar phosphates, which may bind to either the carbamoylated (i.e., CA1P; refs. 4 and 5) or uncarbamoylated (i.e., RuBP; ref. 6) forms, and thus it can maintain Rubisco in an almost fully active form even at otherwise limiting CO2 concentrations (7, 8). ADP is an inhibitor of activase activity, and under typical physiological conditions, activase exhibits half or less of its maximal activity in vitro (9).
In Arabidopsis thaliana and other species, the activity of Rubisco varies with light intensity over the same range as photosynthesis by means of changes in the relative amounts of active and inactive forms of the enzyme (10, 11). However, the stromal ADP/ATP ratio under steady-state conditions does not vary greatly with light intensity (12, 13), and a signaling mechanism for the change in Rubisco activity with light intensity has remained obscure. Recently, redox changes in the larger isoform that are mediated by thioredoxin-f were shown to regulate the activity of the activase in vitro (9). Redox alters the sensitivity of the activase to inhibition by ADP over the physiological range of ATP/ATP observed in the chloroplast stroma. Thus, a signaling pathway involving redox changes in the stroma that modulate the activity of the activase might account for light dependence of the Rubisco activation state observed in vivo. However, there is little direct evidence in support of this mechanism in vivo. There is only one report of a change in the extractable activity of activase, and the dependence on light intensity saturated before that of Rubisco itself (14). Also in some species, such as tobacco, the larger isoform of activase is not present, yet a light intensity dependence of the Rubisco activation state in tobacco is still observed (15).
The Arabidopsis rca mutant does not express either isoform of activase because of a point mutation that disrupts the alternative splicing process (16). Here we have transformed this mutant with a variety of Arabidopsis activase cDNAs under the control of a strong constitutive promoter and examined their Rubisco activation characteristics. Transformants were identified that express the small isoform of activase, the larger isoform, both isoforms by means of a genetic cross, or a larger isoform lacking one or both of the cysteine residues required for redox regulation. This strategy enabled us to resolve the role of each isoform and redox regulation in the activation and light modulation of Rubisco in Arabidopsis in vivo.
Materials and Methods
Gene Constructs for Plant Expression.
The cDNA of the 43-kDa isoform of Rubisco activase from the pTrc99a expression vector (17) was excised and cloned into PstI sites of the pCGN1547 (Calgene, Davis, CA) vector, and the resulting construct was named pCGN1547–43. The cDNAs of the 46-kDa isoform, the 46-kDa C411A mutant, and the 46-kDa C411A+C392A double-mutant isoforms (9) were cloned into BamHI/SalI sites of a pBIN121 (CLONTECH) vector that was altered to contain these sites in front of the nopaline synthase terminator, to create the pBIN46, pBINCys-5 and pBINDM constructs, respectively. The cauliflower mosaic virus 35S promoter in these constructs results in the constitutive expression of these four cDNAs in the transformed plants.
Plant Transformation and Selection.
pBIN46, pBINCys-5, and pBINDM were separately transferred to Agrobacterium tumefaciens strain GV3101 by triparental mating (18). PCGN1547-43 was transferred to Agrobacterium tumefaciens strain EHA101 by freeze-thaw (19). These Agrobacterium strains were then used to transform Arabidopsis rca plants by in planta vacuum infiltration (20). The primary generation transformed seeds were selected on plates containing Murashige and Skoog basal salts (4.3 g/liter), 1% (wt/vol) sucrose, 0.8% (wt/vol) agar, and 30 μg/liter kanamycin. Plants were selfed, and selection was continued for several generations to obtain homozygosity. Genome Southern blots were performed on these transgenic lines until 1–3 independent transformants with a single insertion were obtained for each construct. One line of each desired transformant was chosen for detailed analysis. To obtain transformed plants expressing both isoforms from the introduced genes, a 43-kDa line was genetically crossed with a 46-kDa line. Individual plants of the first-generation progeny were screened for expression of both isoforms by SDS gel electrophoresis and Western blotting before further experiments were performed.
Plant Growth Conditions.
Wild-type and transgenic Arabidopsis rca plants were grown in a commercial soil (Sunshine Mix no. 1, J. R. Johnson Supply, St. Paul) in a growth chamber at 200 μE⋅m−2⋅s−1 [1 E (einstein) = 1 mol of photons] under fluorescent lights with incandescent supplement over either a 10- or 12-h day/night cycle. For gas exchange measurements, plants were grown in 5-inch (13-cm) pots to obtain large leaves. The rca plants, which cannot survive under normal atmospheric conditions, were grown with 0.2% CO2 in the atmosphere under the same conditions.
Measurement of Activase Content.
The amount of activase in the plants was measured by chemiluminescent detection on immunoblots after separation of the soluble protein in leaf extracts by SDS gel electrophoresis as described (21).
Photosynthesis Measurements.
CO2 assimilation and the calculated internal partial pressure of CO2 were measured with a LI-COR 6400 (LI-COR, Lincoln, NE) portable photosynthesis system at 600 μE⋅m−2⋅s−1, a leaf temperature of 23°C, and in the presence of 21% O2. Internal CO2 was varied with the CO2-injection system in the instrument. Measurements were made on individual leaves ≈7–8 weeks after planting. Leaf area in the chamber was determined after completion of the measurements with diazo paper, and area analysis of the resulting images was performed with Image Tool Software (Univ. of Texas Health Science Center, San Antonio).
Activation State and RuBP Measurements in Light Transition Experiments.
Plants grown for approximately 4 weeks were taken to the laboratory (room levels of 390–400 μbar of CO2) and exposed to a light intensity of 600 μE⋅m−2⋅s−1 for 60 min before leaves were removed for the first sample point. This step was eliminated for the rca plants. Afterward, the light intensity was reduced to 35 μE⋅m−2⋅s−1, and leaves were removed after 15, 30, and 60 min of exposure. Finally, more leaves were collected after a return of the light intensity to 600 μE⋅m−2⋅s−1 for 15, 30, and 60 min. Immediately after removal, the leaves were frozen in liquid nitrogen and stored at −80°C. The frozen leaves were ground into a fine powder, and 0.5 ml of extraction buffer containing 100 mM N-tris(hydroxymethyl)glycine (Tricine)⋅KOH (pH 8.0), 1 mM EDTA, 2 mM DTT, 10 μM leupeptin sulfate, and 1 mM PMSF was added. After rapid mixing, the sample was centrifuged for 10 s at 10,000 × g. The initial Rubisco activity was assayed by adding 50 μl of supernatant into 450 μl of assay buffer containing 100 mM Tricine⋅KOH (pH 8.0), 10 mM NaHCO3, 0.5 mM RuBP, and 10 mM NaH14CO3 (2 μCi/μmol; 1 Ci = 37 GBq). The reaction was terminated after 30 s by adding 150 μl of 4 M formic acid/1 M HCl. The total Rubisco activity was assayed by first adding 40 μl of a mixture containing 150 mM NaHCO3 and 150 mM MgCl2 to 360 μl of the supernatant, and then by incubation of the sample at room temperature for 10 min. Subsequently, 50 μl of this sample was added to 450 μl of assay buffer, and the reaction was terminated as described above. The samples were dried overnight at 70°C, and the incorporation of 14CO2 into acid-stable products was determined by liquid scintillation counting. RuBP was measured by addition of 0.55 ml of 100 mM Tricine (pH 8.0) and 15 mM EDTA to the powered leaf samples. After rapid mixing, the sample was centrifuged for 30 s at 10,000 × g, and 0.45 ml of the supernatant was added to 23 μl of 70% perchloric acid; the remainder was saved for assay of protein. After storage on ice for 5 min, the resulting precipitate was removed by centrifugation, and 0.425 ml of the supernatant was neutralized to pH 8 by addition of 4 M KOH, and 1 M MgCl2 was added to reach 25 mM. The resulting precipitate was again removed by centrifugation. The amount of RuBP in 0.4 ml of the supernatant was measured by the addition of 0.2 ml of assay buffer containing 100 mM Tricine (pH 8.0), 25 mM MgCl2, 60 mM NaH14CO3 (0.2 μCi/μmol), and 50 μg of Rubisco. After 30 min at room temperature, reactions were terminated, and incorporation of 14CO2 was determined as described above. Recovery and completeness of the assay were verified in control experiments by addition of known amounts of RuBP to leaf extracts. The content of Rubisco-binding sites in the plants was measured by using 2-[14C]carboxyarabinitol 1,5-bisphosphate as described (21).
Fructose Bisphosphatase Activity Measurements in Light Transition Experiments.
Fructose bisphosphatase activity in soluble protein extracts of the leaves was measured with a spectrophotometric assay as described (22).
Miscellaneous Procedures.
DNA cloning and PCR techniques followed standard procedures (23). The Biotechnology Center at the University of Illinois performed all DNA sequencing.
Results
Transformation of the Arabidopsis rca mutant, which does not express activase, provided an ideal means to examine the function of each activase isoform independent of the other. Transformation of Arabidopsis by vacuum infiltration of transformed Agrobacterium is known to be efficient (24). In our case, after selection for homozygosity and a single-copy insertion by Southern blot genomic analysis, one transgenic line was usually obtained from each plant subjected to vacuum infiltration. Complementation of the rca mutant with either the 43- or the 46-kDa isoform of activase restored the ability of the plants to grow under normal levels of CO2.
The cauliflower mosaic virus 35S promoter used in our constructs typically results in high expression. Activase expression levels were determined by immunoblot analysis after separation of the soluble proteins in leaf extracts by SDS gel electrophoresis (Fig. 1). In the wild-type plants, the two activase isoforms were expressed at a 1:1 ratio, and together they constituted 5% of the total soluble protein, whereas activase protein was undetectable in the rca mutant plants. These results are consistent with a recent report (21). Compared with the wild type, activase expression was lower in both the 43-kDa line (71%) and the 46-kDa line (51%). However, on an individual basis, expression in the 43-kDa line was actually greater (+39%), and expression in the 46-kDa line was about the same as in the wild type.
Figure 1.
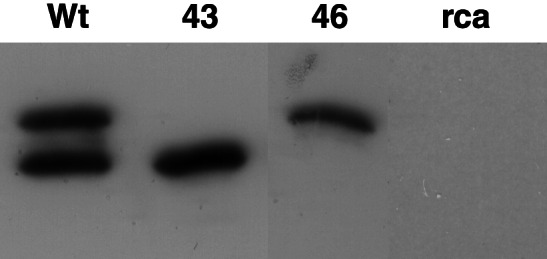
Activase expression in leaves of the wild type (Wt), rca mutant (rca), and rca transgenics expressing the 43-kDa (43) or 46-kDa (46) isoforms. A typical result is shown in which each lane contained 2 μg of total soluble protein. The figure is a composite created by eliminating intervening lanes on the original immunoblot and the addition of the “rca” lane from a different immunoblot obtained under identical conditions. Immunoblot quantitation by densitometry with recombinant standards indicated that expression in the wild type was 110 mg⋅m−2 at nearly a 1:1 ratio (51% vs. 49%). Expression as compared with 100% for the wild type averaged 71% ± 12% in the 43-kDa plants and 51% ± 8% in the 46-kDa plants (n ≥ 4).
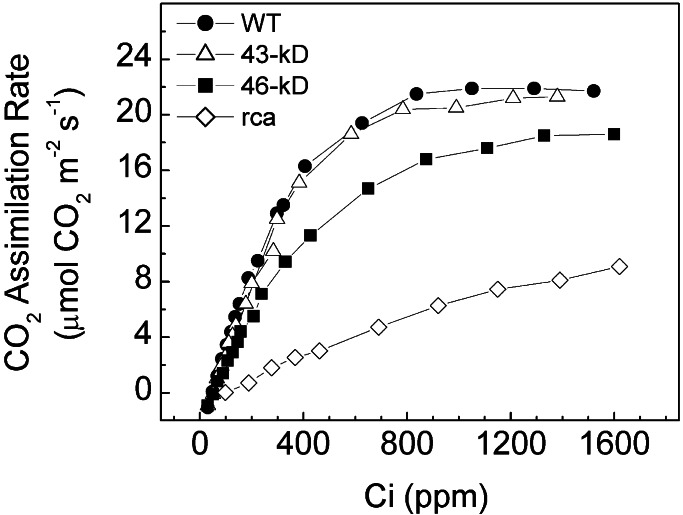
Studies in which activase expression in both Arabidopsis and tobacco was reduced by transformation with an antisense construct (5, 21, 25) indicated that large reductions in activase levels are required before steady-state photosynthesis is noticeably affected. Also, in vitro experiments with the recombinant isoforms indicate that the 46-kDa isoform does not activate Rubisco as effectively as does the 43-kDa isoform (9, 17). To determine whether the level of activase expression or the type of isoform in the transformants had any adverse effects on steady-state photosynthesis by the transformants, we measured the dependence of CO2 assimilation on the intracellular concentration of CO2 (Fig. 2). As reported (26), the rca plants exhibited a very low assimilation rate, even at elevated CO2 concentrations as compared with the wild-type plants. The response of the 43-kDa transformants was nearly identical to the wild-type plants, whereas both the initial slope and maximal rate were significantly lower in the 46-kDa transformants.
Figure 2.
Dependence of the CO2 assimilation rate on the intracellular CO2 concentration (Ci) in the wild type (●), rca mutant (⋄), and rca transformants expressing either the 43-kDa (▵) or 46-kDa (□) isoform of activase. The calculated initial slopes were as follows: WT, 0.061; 43-kDa, 0.054; 46-kDa, 0.040; and rca, 0.01.
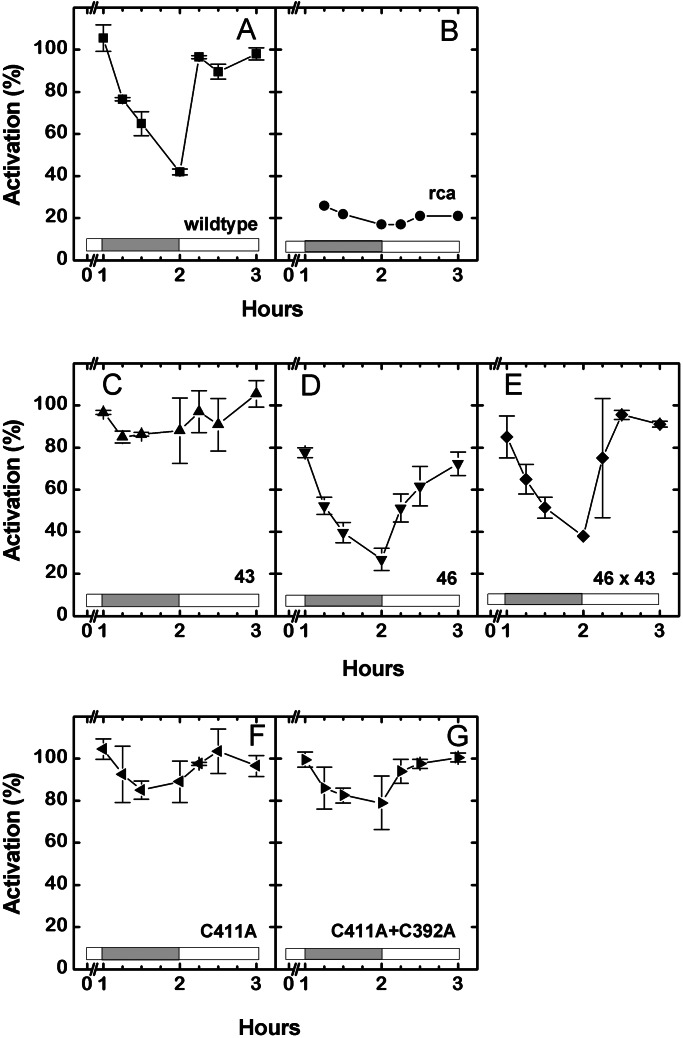
Previous experiments on the wild-type plants have shown that the activation state of Rubisco under normal atmospheric conditions varies with light intensity, ranging from about 40% maximal at very low light to nearly 100% at high light (11). In sharp contrast, the activation state in the rca plants decreases on illumination with either high or low light intensities (27). To compare Rubisco activation in the transformants with the wild-type and the rca plants, we measured the activation state of Rubisco at various times during the exposure of plants to a high–low–high light regime. As shown in Fig. 3A, the activation state in the wild-type plants slowly decreases over the course of 1 h from 100% to 40% after the transition from high to low light. Additional experiments (not shown) indicate the activation level does not decrease much further with continued exposure to low light. Re-exposure to high light results in a rapid (within 15 min) and complete reactivation of Rubisco. Rubisco activation in the rca plants was very low, remaining about 20% of maximal throughout the experiment (Fig. 3B). In separate experiments, RuBP levels in the wild-type plants were also measured. In agreement with previous reports examining Arabidopsis (11) and other species (reviewed in ref. 28), the level of RuBP was initially in excess of the level of Rubisco-binding sites under high light. After the transition to low light, RuBP initially decreased and then increased to approach the starting level after continued exposure to low light (Table 1), reflecting the slow decrease in Rubisco activation. The RuBP level in the rca plants was much higher than in the wild-type plants, in agreement with previous observations (27). RuBP also decreased after the light was reduced in the rca plants, but remained more than 2-fold greater than the Rubisco binding site level, reflecting the very low activation of Rubisco in these plants at both high and low light intensity.
Figure 3.
Time course of Rubisco activation during a transition from high light to low light for 1 h and a return to high light in various plants. The plants examined are the wild type (A), the rca mutant (B), 43-kDa isoform transformant of the rca mutant (C), 46-kDa isoform transformant of the rca mutant (D), progeny of a cross between the 43- and 46-kDa transformants (E), 46-kDa isoform transformant with a C411A substitution (F), and 46-kDa isoform transformant with C411Aand C392A substitutions (G). Data are the means of two or three replicates.
Table 1.
RuBP levels in wild-type and transgenic plants
| Plant | RuBP, nmol
per mg of protein
|
||
|---|---|---|---|
| High light | 15 min of low light | 60 min of low light | |
| Exp. 1 | |||
| Wild type | 13.0 ± 1.7 | 5.7 ± 0.8 | 9.9 ± 1.6 |
| 43-kDa | 14.1 ± 4.5 | 2.0 ± 1.0 | 1.4 ± 0.3 |
| Exp. 2 | |||
| Wild type | 7.8 ± 0.5 | 1.4 ± 0.1 | 3.2 ± 0.8 |
| 43-kDa | 7.4 ± 1.2 | 1.1 ± 0.4 | 0.7 ± 0.3 |
| 46-kDa | 23.4 ± 4.6 | 5.0 ± 3.0 | 14.2 ± 4.1 |
| rca | 28.7 ± 4.0 | 10.1 ± 3.7 | 18.9 ± 5.2 |
Rubisco binding site level was 4.5 ± 0.3 nmol per mg of protein. Data are means and SE for three or more replicates.
In marked contrast to both the wild type and the rca mutant, the activation state of Rubisco in rca transformants expressing only the 43-kDa isoform of activase remained above 80% throughout the low light exposure (Fig. 3C). Thus, Rubisco activation in these plants no longer decreases at low light intensities. Additional experiments revealed that RuBP levels in these plants were similar to those in the wild type, being in excess of the Rubisco active sites under high light (Table 1). But unlike the wild type, after the initial decrease on exposure to low light, the RuBP levels in the 43-kDa transformant did not increase with continued exposure to low light, consistent with the maintenance of a high activation state.
The activation state characteristics of Rubisco in the transformants of rca expressing only the 46-kDa isoform (Fig. 3D) were more similar to, yet clearly distinguishable from, the wild-type plants. The initial activation state in the 46-kDa transformant under high light was about 20% lower than that exhibited by the wild-type plants. Exposure to low light for 1 h lowered the activation state at the same rate and by about the same amount as in the wild type, to reach a limiting value of only 20%. However, the reactivation of Rubisco in the 46-kDa transformants after the plants were returned to high light took about 1 h compared with the 15 min for the wild-type plants. Similar to the wild-type plants, the RuBP level in these transformants initially decreased but then increased with continued exposure to low light (Table 1). The results in Fig. 3 A–D clearly show that the 46-kDa isoform of activase is required for the modulation of Rubisco activity by light intensity.
As mentioned above, the dramatic differences in the Rubisco activation time course in the 43- and 46-kDa transformants are consistent with the ability of the recombinant isoforms to activate Rubisco in vitro. Furthermore, experiments with the recombinant isoforms showed that the activity of the smaller (43-kDa) isoform could be controlled indirectly by redox changes in the presence of the larger isoform (9, 29). To demonstrate this control in planta and to further demonstrate that the differences in Rubisco activation were due only to the type of isoform reintroduced into the rca plants, we crossed the 46- and 43-kDa transformants. We then examined the Rubisco activation characteristics of progeny expressing both isoforms (Fig. 3E). The response of Rubisco activation in these plants to the high–low–high light transitions was nearly identical to that observed in the wild-type plants, demonstrating that the transgenic larger isoform was able to control the activity of the transgenic smaller isoform in planta even when they were expressed from different genes.
The ability of reduction/oxidation (redox) to regulate the activity of native Rubisco activase in vitro has been attributed to a C-terminal extension unique to the larger isoform. This domain contains two Cys residues, and the replacement of either with Ala abolished the ability of redox changes mediated by thioredoxin-f to alter the activity of the recombinant larger isoform (9). The results of these experiments and the activation characteristics of the 43- and 46-kDa activase transformants suggested that, in planta, the response of the Rubisco activation state to light intensity could be mediated by redox changes in the chloroplast stroma. To further test this hypothesis, we examined the Rubisco activation characteristics of 46-kDa transformants in which Cys-411 or both Cys-411 and Cys-392 residues in the C-terminal portion of the larger isoform were replaced with Ala. As shown in Fig. 3 F and G, Rubisco activation remained high in these both transformants during the low light exposure, quite similar to the response of the 43-kDa transformant (Fig. 3C). These results show that these two Cys residues in the C-terminal extension are crucial for the light modulation of Rubisco activity in planta.
The responses of the fructose bisphosphatase and Rubisco to light intensity are very similar (22). To demonstrate that the change in the light regulation of Rubisco in the transformed lines was specific to the redox state of the 46-kDa isoform of activase, the activity of fructose bisphosphatase in the wild-type, 43-kDa, 46-kDa, and C4111A lines was measured in an experiment using the same light transition protocol as was used for Rubisco activation state measurements. During the high light treatment at both the beginning and end of the experiment, the extractable fructose bisphosphatase activity was about 0.3 nmol⋅min−1⋅μg−1 soluble protein in all plants. The fructose bisphosphatase activity declined by about 75% in all plants after 15 min of low light exposure and remained at this level for the next 45 min (data not shown). Thus the stromal ferredoxin–thioredoxin regulatory system that regulates fructose bisphosphatase was not disrupted in our transformants.
Discussion
In the results reported here, we used transformed plants expressing various forms of the activase to show that the larger isoform of Arabidopsis Rubisco activase plays a unique role in the regulation of Rubisco in planta. Plants not expressing the larger isoform or with Cys→Ala substitutions in the C-terminal extension unique to this isoform lost the capacity to down-regulate Rubisco when exposed to low light intensities. Thus, redox regulation of the activase is required for the down-regulation process to occur. The results are consistent with expectations arising from a previous study of the Rubisco activation and ATPase activities of the two recombinant Arabidopsis isoforms and Cys→Ala mutation in the C-terminal extension of the larger isoform (9). In that study, redox changes mediated by thioredoxin-f were shown to alter the response of the activase to inhibition by the ADP/ATP ratio in the stroma. However, with in vitro experiments, one could only infer that redox regulation of the larger isoform might be critical for the light intensity dependence of the Rubisco activation state. In the present study with transgenic plants, it is quite clear that plants unable to redox-regulate the activase are incapable of down-regulating Rubisco under limiting light conditions. As a consequence, RuBP is not maintained above the Rubisco active-site level under steady-state conditions with limiting light intensities, and therefore RuBP concentration, not the amount of active enzyme, limits flux through the carboxylation step.
The question of how Rubisco is regulated by light in species like tobacco, which contain only the smaller isoform, remains an enigma. Although it is possible that an entirely different mechanism has evolved for regulating Rubisco or the activase itself, redox sensing remains the best explanation for the reported observations. We speculate that redox regulation can be mediated by another protein that contains a domain homologous to the C terminus of the larger isoform. A recent blast search did not uncover any candidate proteins in tobacco or other species, but curiously a hypothetical Arabidopsis protein (MSJ11.24) does have a homologous C-terminal domain (42% identical) with two conserved Cys and a putative chloroplast targeting sequence.
To our knowledge, the transgenic plants expressing the 43-kDa isoform or the 46-kDa isoform with Cys substitutions that were identified for this study are among the first examples in which the deregulation of a Calvin cycle enzyme in a plant has been achieved via elimination of the redox regulation capacity at a target enzyme. The physiological importance of redox regulation of target enzymes by the ferredoxin–thioredoxin system in plants remains unclear (30), largely because mutants in the regulatory system have not yet been reported. However, plants in which the redox regulation of sedoheptulose bisphosphatase has been partially eliminated are currently being investigated (C. A. Raines and S. Shigeoka, personal communications).
It has been proposed that redox regulation mainly acts as an on/off switch to prevent the futile cycling of Calvin cycle intermediates in the dark, which wastes energy (31). In the case of Rubisco, maximal catalytic potential in the dark would be of little apparent consequence as long as the preceding enzyme, phosphoribulokinase, which is also regulated by redox, remained inactive in the dark and did not provide any RuBP substrate. However, the in vitro studies with the recombinant activase isoforms (9, 17) suggest that the maintenance of the high Rubisco activation state in the dark in the 43-kDa and the Cys-substituted 46-kDa transgenic plants may be at the expense of a high rate of energy consumption by the ATPase activity of the activase. Thus, although futile cycling is not a concern, unnecessary energy consumption in the dark may compromise the growth potential of the 43-kDa transgenic plants. On the other hand, the maintenance of Rubisco at full capacity in the 43-kDa transformants, able to respond instantly to changes in RuBP concentration, could result in more efficient photosynthesis under some circumstances, such as fluctuating light environments.
A more elaborate role for redox regulation may be that it also serves to modulate the activities of the target enzymes in response to changes in the light environment, providing one means for homeostatic control of Calvin cycle activity (e.g., ref. 32). Evidence that the activities of Rubisco and fructose bisphosphatase both vary over a similar range of light intensities as overall photosynthesis (22) provides strong support for this concept. Thus, the impact of deregulating only one enzyme in a system like the Calvin cycle, with multiple sites and layers of control, may be rather limited.
One primary consequence of deregulating Rubisco, as shown in this report, is that RuBP levels do not return to the high levels typical of the wild-type plants after an extended period at low light. The significance of RuBP concentrations in excess of Rubisco active sites in wild-type plants remains obscure (28), but a recent model (8) suggests that it plays a role in the regulation of the carbamoylation state by the activase. Also, RuBP normally constitutes a very large fraction of the total stromal phosphate pool, and thus, in the deregulated transformants, as-yet-uncovered compensating adjustments in chloroplast metabolism may be required. Casual observations indicate that the dramatic change in the regulation of Rubisco has no gross phenotype, as all of the transformed plants in this study appear to grow well. Further studies comparing the wild-type and 43-kDa transgenic plants should provide clear insights into the importance of maintaining high RuBP concentrations at low light intensities and they clearly have the potential to extend our understanding of the importance of regulating Rubisco in the context of whole plant growth and photosynthesis.
Acknowledgments
This work was supported in part by U.S. Department of Energy Grant DE-AI02-97ER20268.
Abbreviations
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- RuBP
ribulose 1,5-bisphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lorimer G H, Badger M R, Andrews T J. Biochemistry. 1976;15:529–536. doi: 10.1021/bi00648a012. [DOI] [PubMed] [Google Scholar]
- 2.Portis A R., Jr Annu Rev Plant Physiol Plant Mol Biol. 1992;43:415–437. [Google Scholar]
- 3.Salvucci M E, Ogren W L. Photosynth Res. 1996;47:1–11. doi: 10.1007/BF00017748. [DOI] [PubMed] [Google Scholar]
- 4.Robinson S P, Portis A R., Jr FEBS Lett. 1988;233:413–416. [Google Scholar]
- 5.Mate C J, Hudson G S, von Caemmerer S, Evans J R, Andrews T J. Plant Physiol. 1993;102:1119–1128. doi: 10.1104/pp.102.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z Y, Portis A R., Jr Plant Physiol. 1992;99:1348–1353. doi: 10.1104/pp.99.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Portis A R, Jr, Salvucci M E, Ogren W L. Plant Physiol. 1986;82:967–971. doi: 10.1104/pp.82.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mate C J, von Caemmerer S, Evans J R, Hudson G S, Andrews T J. Planta. 1996;198:604–613. doi: 10.1007/BF00262648. [DOI] [PubMed] [Google Scholar]
- 9.Zhang N, Portis A R., Jr Proc Natl Acad Sci USA. 1999;96:9438–9443. doi: 10.1073/pnas.96.16.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perchorowicz J T, Raynes D A, Jensen R G. Proc Natl Acad Sci USA. 1981;78:2985–2989. doi: 10.1073/pnas.78.5.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks A, Portis A R., Jr Plant Physiol. 1988;87:244–249. doi: 10.1104/pp.87.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks A, Portis A R, Jr, Sharkey T D. Plant Physiol. 1988;88:850–853. doi: 10.1104/pp.88.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietz K J, Heber U. Biochim Biophys Acta. 1986;848:392–401. [Google Scholar]
- 14.Lan Y, Woodrow I E, Mott K A. Plant Physiol. 1992;99:304–309. doi: 10.1104/pp.99.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruuska S A, Andrews T J, Badger M R, Price G D, von Caemmerer S. Plant Physiol. 2000;122:491–504. doi: 10.1104/pp.122.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orozco B M, McClung C R, Werneke J M, Ogren W L. Plant Physiol. 1993;102:227–232. doi: 10.1104/pp.102.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallis R P, Ewy R G, Portis A R., Jr Plant Physiol. 2000;123:1077–1086. doi: 10.1104/pp.123.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogers S G, Horsch R B, Fraley R T. Methods Enzymol. 1986;118:627–640. [Google Scholar]
- 19.Holster M. Mol Gen Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 20.Bechtold N, Pelletier G. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 21.Eckardt N A, Snyder G W, Portis A R, Jr, Ogren W L. Plant Physiol. 1997;113:575–586. doi: 10.1104/pp.113.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassenrath-Cole G F, Pearcy R W, Steinmaus S. Photosynth Res. 1994;41:295–302. doi: 10.1007/BF00019407. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Bent A F. Plant Physiol. 2000;124:1540–1547. doi: 10.1104/pp.124.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond E T, Andrews T J, Mott K A, Woodrow I E. Plant J. 1998;14:101–110. doi: 10.1046/j.1365-313X.1998.00103.x. [DOI] [PubMed] [Google Scholar]
- 26.Somerville C R, Portis A R, Jr, Ogren W L. Plant Physiol. 1982;70:381–387. doi: 10.1104/pp.70.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvucci M E, Portis A R, Jr, Ogren W L. Plant Physiol. 1986;80:655–659. doi: 10.1104/pp.80.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Caemmerer S, Quick W P. In: Photosynthesis: Physiology and Mechanism. Leegood R C, Sharkey T D, von Caemmerer S, editors. Dordrecht, The Netherlands: Kluwer; 2000. pp. 85–113. [Google Scholar]
- 29.Zhang N, Schürmann P, Portis A R., Jr Photosynth Res. 2001;68:29–37. doi: 10.1023/A:1011845506196. [DOI] [PubMed] [Google Scholar]
- 30.Jacquot J P, Lancelin J M, Meyer Y. New Phytol. 1997;136:543–570. doi: 10.1046/j.1469-8137.1997.00784.x. [DOI] [PubMed] [Google Scholar]
- 31.Buchanan B B. Arch Biochem Biophys. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- 32.Fridlyand L E, Backhausen J E, Scheibe R. Photosynth Res. 1999;61:227–239. [Google Scholar]