Abstract
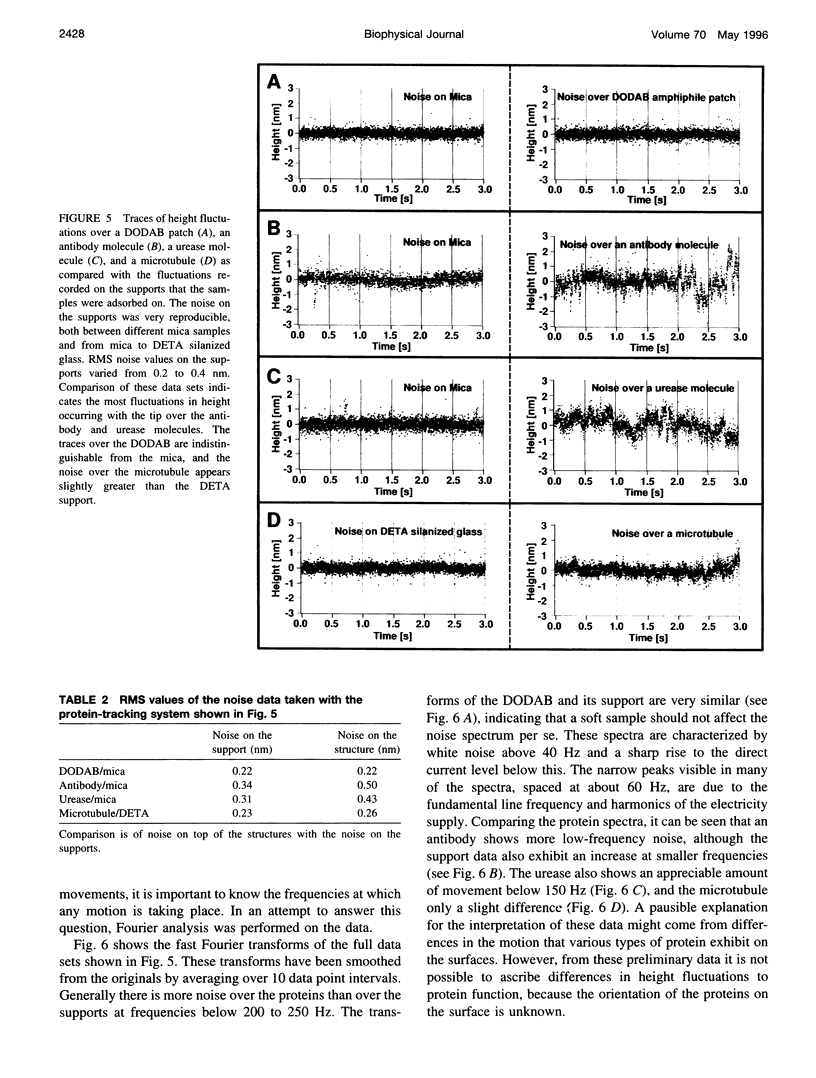
Height fluctuations over three different proteins, immunoglobulin G, urease, and microtubules, have been measured using an atomic force microscope (AFM) operating in fluid tapping mode. This was achieved by using a protein-tracking system, where the AFM tip was periodically repositioned above a single protein molecule (or structure) as thermal drifting occurred. Height (z-piezo signal) data were taken in 1 - or 2-s time slices with the tip over the molecule and compared to data taken on the support. The measured fluctuations were consistently higher when the tip was positioned over the protein, as opposed to the support the protein was adsorbed on. Similar measurements over patches of an amphiphile, where the noise was identical to that on the support, suggest that the noise increase is due to some intrinsic property of proteins and is not a result of different tip-sample interactions over soft samples. The orientation of the adsorbed proteins in these preliminary studies was not known; thus it was not possible to make correlations between the observed motion and specific protein structure or protein function beyond noting that the observed height fluctuations were greater for an antibody (anti-bovine IgG) and an enzyme (urease) than for microtubules.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bezanilla M., Drake B., Nudler E., Kashlev M., Hansma P. K., Hansma H. G. Motion and enzymatic degradation of DNA in the atomic force microscope. Biophys J. 1994 Dec;67(6):2454–2459. doi: 10.1016/S0006-3495(94)80733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986 Mar 3;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Bustamante C., Vesenka J., Tang C. L., Rees W., Guthold M., Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry. 1992 Jan 14;31(1):22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- Dye R. B., Fink S. P., Williams R. C., Jr Taxol-induced flexibility of microtubules and its reversal by MAP-2 and Tau. J Biol Chem. 1993 Apr 5;268(10):6847–6850. [PubMed] [Google Scholar]
- Florin E. L., Moy V. T., Gaub H. E. Adhesion forces between individual ligand-receptor pairs. Science. 1994 Apr 15;264(5157):415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Sligar S. G., Wolynes P. G. The energy landscapes and motions of proteins. Science. 1991 Dec 13;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- Han W., Mou J., Sheng J., Yang J., Shao Z. Cryo atomic force microscopy: a new approach for biological imaging at high resolution. Biochemistry. 1995 Jul 4;34(26):8215–8220. doi: 10.1021/bi00026a001. [DOI] [PubMed] [Google Scholar]
- Hansma H. G., Hoh J. H. Biomolecular imaging with the atomic force microscope. Annu Rev Biophys Biomol Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- Jabri E., Carr M. B., Hausinger R. P., Karplus P. A. The crystal structure of urease from Klebsiella aerogenes. Science. 1995 May 19;268(5213):998–1004. [PubMed] [Google Scholar]
- Johnson L. N. Time-resolved protein crystallography. Protein Sci. 1992 Oct;1(10):1237–1243. doi: 10.1002/pro.5560011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrasch S., Hegerl R., Hoh J. H., Baumeister W., Engel A. Atomic force microscopy produces faithful high-resolution images of protein surfaces in an aqueous environment. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):836–838. doi: 10.1073/pnas.91.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. U., Chrisey L. A., Colton R. J. Direct measurement of the forces between complementary strands of DNA. Science. 1994 Nov 4;266(5186):771–773. doi: 10.1126/science.7973628. [DOI] [PubMed] [Google Scholar]
- Petsko G. A., Ringe D. Fluctuations in protein structure from X-ray diffraction. Annu Rev Biophys Bioeng. 1984;13:331–371. doi: 10.1146/annurev.bb.13.060184.001555. [DOI] [PubMed] [Google Scholar]
- Radmacher M., Fritz M., Hansma H. G., Hansma P. K. Direct observation of enzyme activity with the atomic force microscope. Science. 1994 Sep 9;265(5178):1577–1579. doi: 10.1126/science.8079171. [DOI] [PubMed] [Google Scholar]
- Schabert F. A., Henn C., Engel A. Native Escherichia coli OmpF porin surfaces probed by atomic force microscopy. Science. 1995 Apr 7;268(5207):92–94. doi: 10.1126/science.7701347. [DOI] [PubMed] [Google Scholar]
- Steinbach P. J., Ansari A., Berendzen J., Braunstein D., Chu K., Cowen B. R., Ehrenstein D., Frauenfelder H., Johnson J. B., Lamb D. C. Ligand binding to heme proteins: connection between dynamics and function. Biochemistry. 1991 Apr 23;30(16):3988–4001. doi: 10.1021/bi00230a026. [DOI] [PubMed] [Google Scholar]
- Venier P., Maggs A. C., Carlier M. F., Pantaloni D. Analysis of microtubule rigidity using hydrodynamic flow and thermal fluctuations. J Biol Chem. 1994 May 6;269(18):13353–13360. [PubMed] [Google Scholar]
- Williams R. C., Jr, Lee J. C. Preparation of tubulin from brain. Methods Enzymol. 1982;85(Pt B):376–385. doi: 10.1016/0076-6879(82)85038-6. [DOI] [PubMed] [Google Scholar]
- Yang J., Mou J., Shao Z. Molecular resolution atomic force microscopy of soluble proteins in solution. Biochim Biophys Acta. 1994 Mar 2;1199(2):105–114. doi: 10.1016/0304-4165(94)90104-x. [DOI] [PubMed] [Google Scholar]






