Abstract
Otitis media (OM) is the most common childhood disease. Almost all children experience at least one episode, but morbidity is greatest in children who experience chronic/recurrent OM (COME/ROM). There is mounting evidence that COME/ROM clusters in families and exhibits substantial heritability. Subjects who had tympanostomy tube surgery for COME/ROM (probands) and their families were recruited for the present study, and an ear examination was performed, without knowledge of the subject’s history, to determine presence of OM sequelae. In addition, tympanometric testing was performed at three frequencies (226, 630 or 710, and 1,400 Hz) to detect abnormal middle-ear mechanics, and hearing was screened at 20 dB for the speech frequencies. Of these families, 121 had at least two individuals who had received the diagnosis of COME/ROM (364 affected and genotyped individuals), of whom 238 affected and informative relative pairs were used for analyses. Single-point nonparametric linkage analysis provided evidence of linkage of COME/ROM to chromosome 10q at marker D10S212 (LOD 3.78; P=3.0×10-5) and to chromosome 19q at marker D19S254 (LOD 2.61; P=5.3×10-4). Analyses conditional on support for linkage at chromosomes 10q and 19q resulted in a significant increase in LOD score support on chromosome 3p (between markers D3S4545 and D3S1259). These results suggest that risk of COME/ROM is determined by interactions between genes that reside in several candidate regions of the genome and are probably modulated by other environmental risk factors.
Introduction
Otitis media (OM) is the most common childhood disease. Almost all children experience at least one episode, but morbidity is greatest in children who experience chronic/recurrent OM (COME/ROM). There is mounting evidence that COME/ROM clusters in families and exhibits substantial heritability. Several studies have reported a greater risk of COME/ROM in children with a family history of middle-ear disease, which suggests a genetic component. However, development of COME/ROM among siblings may be due to inherited biologic susceptibility, common environmental exposures, or an interaction between them. Prospective studies have shown that children whose siblings have a history of COME/ROM have a 1.6–4.2-fold increase in risk, after controlling for environmental factors (Sipila et al. 1988; Teele et al. 1989; Rasmussen 1993). Three twin studies have reported similar heritability estimates for OM. A Norwegian population-based sample of 2,750 twin pairs reported recurrent OM (ROM) heritability of 0.45 and 0.74, respectively, for male and female pairs ascertained by self-report (Kvaerner et al. 1997). Pittsburgh researchers enrolled 175 same-sex twin or triplet sets and followed them prospectively, with regular examinations from birth to age 24 mo (Casselbrant et al. 1999). Heritability for the mean proportion of time with otitis media with effusion (OME) in the first 2 years of life was 0.73; discordance for ROM was 0.37 for DZ and 0.04 for MZ pairs. A prospective study of 715 MZ and 658 DZ twin pairs in the United Kingdom examined heritability of OM symptoms at ages 2, 3, and 4 years (Rovers et al. 2002). In the U.K. study, heritability was estimated as 0.57 for acute OM and as 0.72 for chronic airway blockage. Correlation between the total symptom score at age 2 years was 0.90 for MZ twins and 0.65 for DZ twins.
Although there is evidence that OM has a significant genetic component, no genome screens of families with affected relative pairs have been reported (to our knowledge), and only a few investigators have studied potential candidate genes for COME/ROM. Cytokines are attractive candidates because they play a key role in OM pathogenesis; genes that control cytokine production and function would be expected to be important in OM susceptibility. A Finnish study reported an association between IL-1α and recurrent OM in children without a history of allergic rhinitis (Joki-Erkkilä et al. 2002). Another Finnish study reported an association between the surfactant protein A gene locus and susceptibility to severe respiratory syncytial virus (RSV) infection (Löfgren et al. 2002) and recurrent acute OM (Rämet et al. 2001). A possible association has been suggested between surfactant protein D and RSV, which is an important viral cause of OM (Lahti et al. 2002). Invasive disease caused by Streptococcus pneumoniae, the major pathogen implicated in OM, has been linked to genotypes defined by mutations in codons 52, 54, and 57 of the mannose binding lectin (MBL) gene and results in low levels of MBL (Roy et al. 2002). Cases were 2.6 and 2.0 times more likely than controls to be homozygous for those variants in the original and confirmatory studies, respectively. The present study was designed for the investigation of families' genetic linkage between COME/ROM and polymorphic DNA markers at selected sites across the entire human genome, by use of affected relative pairs.
Material and Methods
Sample Recruitment
Probands (defined as “individuals who had tympanostomy tube surgery for COME/ROM”) and their families were recruited for the present study. In 2002, we began identifying probands from past and current studies conducted at the University of Minnesota Otitis Media Research Center between 1992 and 2001 (129 families) (Daly et al. 1996, 1999; Hunter et al. 1996; Ho et al. 2002) and from the general public (24 families). Of the 129 families recruited from previous studies, 82% of probands were ascertained from studies in which enrollment occurred at the time of tympanostomy tube treatment, 10% were from a prospective study of infants treated with tubes, and 8% were identified by a parent as having had tube treatment in a cross-sectional hearing-screening study. Of the other 24 families, 67% responded to posted advertisements, and 33% were recruited from the Children’s Ear and Hearing Clinic at the University of Minnesota. Ascertainment was broad based across the studies and included children (1) who were treated with tubes at five community hospitals, (2) who received health care at 17 clinics in a health maintenance organization, and (3) who participated in a hearing screening study conducted throughout Minnesota.
Family members were ineligible if they were adopted or if they had Down syndrome, craniofacial anomaly (i.e., cleft palate), another genetic disorder with otologic complications, or tympanostomy tubes for a diagnosis other than OM. The study was reviewed and approved annually by the University of Minnesota and Wake Forest University institutional review boards.
Clinical Evaluation
During a study-specific visit to the University of Minnesota Otolaryngology clinic, family members gave written consent (and, after April 14, 2003, signed a Health Insurance Portability and Accountability Act authorization form). The study neuro-otologist performed an ear examination, without knowledge of the subject’s history, to determine presence of OM sequelae; tympanometric testing was performed at three frequencies (226, 630 or 710, and 1,400 Hz) to detect abnormal middle-ear mechanics; hearing was screened at 20 dB for the speech frequencies; and blood was drawn. Family members provided information, by questionnaire or interview, about their OM history, risk factors, and related conditions and provided medical-record releases. Parents (usually the mothers) provided history information for their children. Trained study personnel abstracted data from the participants' medical records regarding OM history, middle-ear surgery, audiograms, tympanograms, and other contributing medical problems.
Affected-Status Definitions
Criteria for determination of phenotype are depicted in appendix A. An individual was classified as “affected” if (1) at least two different data sources indicated a positive result for the characteristics in appendix A or (2) one data source indicated a positive result and there was a specific middle-ear or tympanometric finding thought by experts to be presumptive evidence of a history of COME/ROM. These findings included tympanosclerosis, atrophy, the presence of a tympanostomy tube, cholesteatoma, static admittance greater than age-referenced norms, and resonant frequency less than the lowest frequency tested.
There were 692 members (688 in the original data, an addition of 5 anonymous “fathers” for creation of half-sib relationships, and a deletion of 1 unrelated subject) in the 133 pedigrees. Of the 692 members, 76 had “unknown” disease status, either through insufficient clinical information or failure to undergo examination. The remaining 616 individuals were considered as either “unaffected” or “affected” on the basis of the classification schemes discussed above. When measured by only the most strict criteria (at least two sources), there were 245 unaffected and 371 affected individuals. Ninety-four percent were white, and 99% were non-Hispanic.
Family Sample
Families were eligible for the linkage study if they had children with concordant and/or discordant histories of COME/ROM and adequate DNA samples. In the original family collection, 153 pedigrees were enumerated. These pedigrees were ascertained from multiple studies and included 72 families from the Family Study of Otitis Media, 34 families from the Long-Term Sequelae of OM Study, 13 families from the Early OM Study, 10 families from the Hearing Screening of Minnesota Children, 16 families from the general public, and 8 families of patients from the Children’s Ear and Hearing Clinic. From the 153 families that had 692 members with clinical data, 652 blood samples and 30 buccal samples were collected. Selection of only those families informative for linkage resulted in 591 DNA samples, from 133 families, that were sent to the Center for Inherited Disease Research (CIDR) for a 9-cM genome screen. These 133 families produced 410 affected relative pairs (162 full-sib pairs, 8 half-sib pairs, 16 grandparent-grandchild pairs, 36 avuncular pairs, 13 first-cousin pairs, 17 unrelated pairs, 3 half-avuncular pairs, and 155 parent-offspring pairs), 238 of whom were informative.
Genome Scan
DNA extraction was performed by the Molecular Genetics Laboratory Core of the University of Minnesota General Clinical Research Center. The CIDR performed a genomewide screen of the 591 samples, by use of 404 fluorescent microsatellite markers. The marker set comprised primarily trinucleotide and tetranucleotide repeat polymorphisms that were based on the Marshfield Genetics version 8 screening set. The average spacing of the markers was 9 cM, and the largest gap in the marker panel between adjacent markers was 20 cM; 238,764 genotypes were generated for the study.
Error Checking
The overall missing-data rate was 3.00%, which reflects 7,444 missing genotypes from a total of 248,219 (study and quality-control samples), with the exclusion of three Y chromosome markers. The error rate was 0.07% per genotype, on the basis of 14 different allele calls from 10,618 paired (quality control) sample events.
Verification of reported familial relationships and marker-error detection was made using PREST (McPeek and Sun 2000) and PedCheck (O’Connell and Weeks 1998) software. Nine families were identified as having segregation problems that were not consistent with reported family structure. Five of those families had a structure that required changing a full-sib to a half-sib relationship. Five families (including one with a full-sib–to–half-sib relationship change) required other structure changes. After these family structure problems were resolved, 236 marker genotypes were designated as “missing.” The 236 marker problems occurred in 233 individuals, with distribution across the genome (no one marker had to be excluded).
NPL Analysis
All genetic analyses were performed on the family data with the multiple classifications of affected status. Since all genetic-analysis results were similar in their support for linkage, with only minor deviations in magnitude of support, we present as affected only those that reported two or more sources of evidence, from the clinical or tympanogram findings.
Single- and multipoint-linkage analyses (based on the consensus map order and distances) were performed using GeneHunter-plus and GeneHunter-NPL software (Kruglyak et al 1996; Kong and Cox 1997; Langefeld et al 2001). Multipoint-linkage analysis was performed using the S(pairs) option of GeneHunter-plus, and maximized LOD scores were calculated under an exponential model, with δ constrained between 0 and 2.
Stratified (conditional) analyses were performed using GeneHunter-plus (Kong and Cox 1997), to examine epistatic effects among the candidate chromosomal regions linked with COME/ROM. To model epistasis (gene-gene interaction), a series of analyses was performed in which each family was assigned a “weight.” The weight assigned to each family, either 0 or 1, for each region was based on the evidence of linkage, as reflected by the maximum-multipoint-likelihood-ratio Z score (Zlr) in that region. To model epistasis, a weight of 1 was assigned to families with a maximum multipoint Zlr>0 at that location, with a corresponding weight of 0 assigned to all others. Only families that received a weight of 1 contributed to the resultant LOD scores. LOD scores that increased by at least 1.18 (corresponding to P=.01) were considered as suggestive for epistasis.
Parametric Linkage Analysis
Parametric LOD scores were calculated, using GeneHunter software (Kruglyak et al 1996), for individual markers on chromosomes 10 and 19. Analyses were performed using a recessive model and a dominant model for transmission of COME/ROM susceptibility. Each model included reduced penetrance for the “at risk” genotypes and sporadic rates for the “not at risk” genotypes. Each model produced the equivalent population prevalence of COME/ROM. Thus, the recessive model had COME/ROM allele frequency of 0.40, with 5% sporadic frequency and 95% penetrance of the “at risk” homozygote. The dominant model had COME/ROM allele frequency of 0.10, with 80% penetrance of the “at risk” genotypes and 5% penetrance of the sporadic cases. Parametric analyses were performed using the heterogeneity parameter, α, that represented the proportion of families with linkage at that specific marker, which resulted in a heterogeneity LOD score (HLOD).
Empirical Significance Levels
Empirical P values for regions of interest were determined by simulation of Mendelian transmission for families, by use of marker allele frequencies observed in the sample. The percentage of LOD scores that were greater than the nominal value, on the basis of simulations (at least 10,000 replicates, depending on the magnitude of the observed LOD), provided the estimated empirical P value. Empirical P values for stratified (conditional) analyses were estimated by permutation of the weight assigned to each family while the number of families that were included in any one analysis were maintained.
Results
Risk-factor characteristics of study families with COME/ROM are summarized in table 1. The agreement between reported OM history and other data sources was examined for subgroups. For parents with a positive history of COME/ROM, agreement with exam was 73% and, with tympanogram, agreement was 61%. Eight percent of parents had missing OM history data. Agreement did not differ by sex, age, or education. For reported sibling OM history, agreement with exam was 65%, agreement with tympanogram was 66%, and agreement with medical records was 73%. Only 1% of siblings had missing OM history data. Among siblings, agreement between data sources did not differ by age, sex, or number of siblings. If OM history was positive, there was at least one other positive data source for 67% of parents and 83% of siblings. If OM history was negative, all other sources were negative for 60% of parents and 53% of siblings.
Table 1.
Characteristics of Study Families
|
No./Total (%) of Family Members with Specified Risk Factor Who Were |
||||
| Risk Factor | Phenotyped(n=616) | Phenotyped andDNA Sampled(n=588) | Affected(n=371) | Unaffected(n=245) |
| Preschool day care | 313/612 (51.1) | 309/585 (52.8) | 237/369 (64.2) | 76/243 (31.3) |
| Exclusively formula fed | 241/581 (41.5) | 234/561 (41.7) | 134/361 (37.1) | 107/220 (48.6) |
| Parental smoking | 302/613 (49.3) | 283/588 (48.1) | 163/370 (44.1) | 139/243 (57.2) |
Single-Point NPL Analysis
Single-point NPL analyses were performed. The NPL Z scores were converted to LOD scores by use of allele-sharing-modeling software (Kong and Cox 1997). This analyses identified seven regions in the genome in which at least one marker had a LOD score >1—on chromosomes 5p, 10q, 12p, 14q, 18q (two locations), and 19q. There was significant evidence of linkage to COME/ROM on chromosome 10q at marker D10S212 (LOD 3.78; P=3.0×10-5). There was also suggestive evidence of linkage to COME/ROM on chromosome 19q at marker D19S254 (LOD 2.61; P=5.3×10-4). No other location exhibited a LOD score >1.17 for any region.
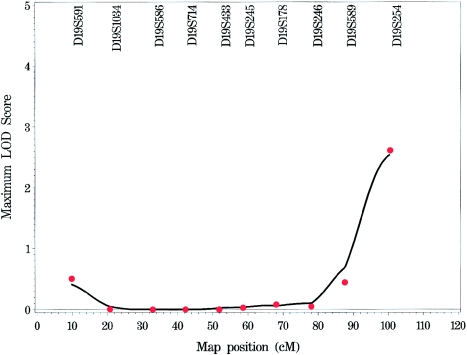
Multipoint NPL Analysis
Multipoint NPL analyses (figs. 1 and A1 [online only]) provided evidence of linkage to COME/ROM similar to that observed in the single-point analyses. The strongest evidence of linkage to COME/ROM occurred on chromosome 19q (LOD 2.53; P=6.4×10-4) near marker D19S254 (fig. 2). This support was similar to that observed in the single-point analysis (LOD 2.61 at marker D19S254). One other location in the genome had a multipoint LOD score >1, on chromosome 10q (LOD 1.64; P=.0064) near marker D10S212 (fig. 3). The evidence that supports linkage on 10q decreased dramatically from that observed in the single-point analysis (LOD 3.78). The observed concordance with respect to location (if not exact LOD score support) between the single-point and the multipoint analyses suggests that the linkage to COME/ROM for these regions may be real and may not represent artifacts generated by genotyping errors.
Figure 1.
Genome scan for COME/ROM
Figure 2.
Linkage analysis of COME/ROM on chromosome 19, with NPL scores for both multipoint (solid line) and single-point (individual points) approaches.
Figure 3.
Linkage analysis of COME/ROM on chromosome 10, with NPL scores for both multipoint (solid line) and single-point (individual points) approaches.
Empirical Significance Levels
We used simulation of genotypes and gene dropping of genotypes under assumed Mendelian transmission to estimate genomewide P values for the observed multipoint-linkage results. The probability of observing a LOD score >1.64 (support on chromosome 10q) by chance was 3.1×10-3 (empirical LOD 1.63). The probability of observing a LOD score >2.53 (support on chromosome 19q) by chance was 3.9×10-4 (empirical LOD 2.46).
Parametric Linkage Analysis
Parametric LOD scores were calculated for markers on chromosomes 10 and 19, since these locations exhibited the strongest evidence of linkage to COME/ROM in both single-point and multipoint analyses. There have been few segregation analyses of COME/ROM in the literature, but the most compelling evidence is that the contribution of a single major gene influences both recessive and dominant modes of inheritance. Analyses were performed both with and without a parameter that allows for heterogeneity of linkage support (with the parameter α representing the proportion of linked families). For chromosome 10, in single-marker analyses, the strongest evidence of linkage was observed for marker D10S212 (HLOD 2.36; α=0.42) under the recessive model and at the same location (HLOD 1.51; α=0.51) under the dominant model. For chromosome 19, the strongest single-point evidence was observed for marker D19S591 (HLOD 1.31; α=0.28) under the recessive model and for marker D19S254 (HLOD 0.70; α=0.32) under the dominant model. It should be noted that the maximum HLOD for chromosome 19 under the recessive model (at marker D19S591) was at the pter location, whereas the maximum HLOD under the dominant model (at marker D19S254) was at the qter location. Differences in magnitude of linkage support between multipoint and two-point analyses could be due to reduced information content of the markers at the telomeric end of 10q (52%).
Conditional Analysis
We performed conditional analyses by stratifying families first into those that supported linkage at 10q and then at 19q and by observing whether regions of the genome exhibited a significant increase in support of linkage for COME/ROM. There was significant overlap in the families that supported linkage at 10q and at 19q, so the regions (if any existed) that had potential interaction effects were expected to be similar. When analyses were performed, a region on 3p (between markers D3S4545 and D3S1259 at 32 cM) exhibited increased support for linkage (unconditional LOD 0.60; conditional [10q] LOD 2.43; conditional [19q] LOD 1.84). It should be noted that D3S1259 is ∼8 cM from D3S3038, a marker that has a single-point HLOD of 1.03 by use of a parametric (recessive) model. Together, these results suggested an interaction between 10q with 3p and between 19q with 3p.
Phenotypic Correlations
There was the appearance of significant overlap in the families that supported linkage of COME/ROM to 10q and 19q. Of the entire set of 133 families, 18 families supported linkage (LOD >0) for only 19q, 25 families supported linkage only for 10q, 38 families supported linkage at both 10q and 19q, and 52 families failed to provide evidence of linkage at either of the two primary locations. There were 36 families that supported both 3p and 10q linkage, and there were 31 families that supported both 3p and 19q linkage; 26 of these families supported linkage to all three locations.
Among all families supporting linkage at one or both of the 10q or 19q locations, 63.0% had more than one family member with a history of tympanostomy tubes (indication of more severe or chronic disease), compared with 36.5% in the families that did not support linkage at these sites (P=.003). The median number of affected relatives in the families supporting linkage was approximately three, compared with two affected relatives in families not supporting linkage (P=.009).
Factors known to be associated with increased risk of COME/ROM in epidemiologic studies were characterized in family members, with respect to evidence of family-linkage (to either 10q or 19q) support. The proportion of family members exposed to exclusive formula feeding was 38.2% in families with linkage, compared with 47.2% in families not supporting linkage (P=.043). The proportion of families with smokers in the household was 48.5%, compared with 51.1% in families that failed to support linkage (P=.585). The proportion of families that employed day care in early childhood was 52.2% in the families with linkage, compared with 49.3% in families not supporting linkage (P=.555).
Discussion
We report the initial genome scan performed for one of the most common chronic disorders in children, COME/ROM. Our sample of families with COME/ROM was enriched with multiple cases, probably those with severity greater than average, and with sufficient evidence that older family members still have evidence of COME/ROM years after the event. However, there is no simple diagnostic test for determination of COME/ROM history. Although younger children are likely to have better agreement between data sources because OM is a disease of early childhood, it is necessary to provide phenotype data for family members of all ages. Affected status was based on data from four sources, to increase the likelihood of accurate phenotyping. All sources are imperfect: history may not be accurately remembered, those with a recurrent OM history are less likely to have definitive changes in the tympanic membrane and middle ear than those who received tubes, and, for older children and (especially) adults, it is a challenge to obtain a pertinent medical record for early childhood. Because of these limitations, some over- and underclassification of affection status may have occurred.
Our results suggest that relatively few genetic locations may be involved in susceptibility to COME/ROM. Overall, we identified several regions that support linkage by use of both single- and multipoint NPL analyses on chromosomes 10q and 19q. When conditional analyses were employed, a potential region on chromosome 3p was observed to interact with those on 10q and 19q. Analyses of these data by parametric methods confirmed the contribution of 10q, provided limited support for an independent effect on 19q, and supported a role for a region on 3p.
The current model for the development of COME/ROM proposes that an initial viral infection is followed by a bacterial infection; progression is dependent on genetically determined anatomical and immunological factors that lead to impairment of ciliary function in the middle ear. Ultimately, there is an establishment of a bacterial biofilm, which may be a factor in the persistence of infection and effusion (Ehrlich and Post 1999). Central to this model is the immunological response to the initial viral infection and to the secondary bacterial persistence and, therefore, to the exacerbation of the inflammatory state. The three candidate regions for OM identified in this study contain plausible candidate genes involved in defense responses or in the modulation of the inflammatory response. Other candidates may be unrecognized, and the support intervals for the regions remain broad and not precisely defined; therefore, the identity of the “true” positional candidates remains unresolved.
The COME/ROM susceptibility region in 19q (19q13.42-q13.43) contains >100 annotated genes (University of California–Santa Cruz [UCSC] Genome Bioinformatics), many of which code for predicted proteins with unknown function. Among the characterized genes, the region includes the ∼1-Mb leukocyte receptor cluster (LRC), which contains the genes encoding 12 killer cell immunoglobulin (IG)-like receptors (KIR) as well as 13 IG-like transcripts (ILTs) (André et al. 2001). KIR proteins negatively regulate the cytotoxic activity of natural killer cells and subsets of T cells, whereas the closely related ILT proteins are expressed in lymphoid and myelomonocytic cells (André et al. 2001). Members of the LRC prevent the activation of immune cells by interaction with specific ligands, such as major histocompatibility complex (MHC) class I molecules. It is noteworthy that, like the MHC, the LRC is highly polymorphic, not only at the level of gene alleles but also in the number of KIR and ILT genes that are encoded in the cluster (Wilson et al. 2000).
The second COME/ROM region on 10q (10q26.3) encodes 11 characterized genes and at least 17 predicted genes (UCSC Genome Bioinformatics). Because of the similarities between the pulmonary and middle-ear epithelia in their response to xenobiotic agents, a plausible candidate gene for COME/ROM in this region is the ADAM8 gene, a member of the ADAM (a disintegrin and metalloproteinase domain) family of proteins that contain the catalytic site of a metalloprotease and an anti-integrin domain (Yamamoto et al. 1999). Recently, ADAM8 was identified as an allergen-induced asthma gene (King et al. 2004) and has been shown to convert membrane-bound CD23 into a soluble form that regulates IgE synthesis, macrophage activation, and inflammation. Although several candidate genes (SFTPA1 and MBL2) have been mapped to 10q, the support interval for linkage on 10q in the present study was at the telomere, quite distinct from the positions of SFTPA1 and MBL2.
The region in 3p25.3 that is putatively involved in a gene-gene interaction with the two previous regions (10q and 19q) includes two interesting candidate genes for COME/ROM—histamine receptor H1 (HRH1) and interleukin-1 (IL-1) receptor activated kinase 2 (IRAK2). HRH1 codes for a G protein–coupled receptor that mediates some of the actions of the inflammatory mediator, histamine (Yamashita et al. 1991). Mice that lack Hrh-1 show suppression of interferon γ and increased secretion of the T helper cytokines IL-4 and IL-13, as well as higher levels of antigen-specific IgG1 and IgE compared with wild-type mice, which suggests that HRH1 modulates the effect of histamine on T cell–mediated immune response (Jutel et al. 2001). IRAK2 is a component of the signal transduction complex that is recruited to the IL-1 and Toll-like family of receptor complexes (Janssens and Beyaert 2003) upon binding of their ligands, which ultimately results in the activation of the NF-κB transcription factor, a known inducer of inflammatory gene expression. It should be recognized, however, that the power of the conditional analyses is less than that of the original linkage analyses, because of the stratification of the families into subsets on the basis of the support for COME/ROM linkage at 10q and 19q. Thus, these results should be viewed with caution.
In summary, these results suggest that the genetic susceptibility to COME/ROM is determined, in part, by the contribution of genes in three distinct chromosomal regions (10q, 19q, and 3p), with potential gene-gene interaction. These regions of linkage contain several putative positional candidate genes, including those involved with inflammation and response to environmental pathogens. Replication of these findings in this and other populations as well as in targeted molecular studies will further refine the genetic cause of this most common chronic disease of childhood.
Acknowledgments
We are grateful to the numerous individuals and families who donated their time and resources. We thank Michelle Bochert, Jennifer Palmer, Kendra Herrell, Judy Monroe, Norrita Rech, Kim Canfield, and Heather Nelson, for support in patient recruitment and collection; Chris Valis and Mindy Cox, for analytic support; and William Oetting and Richard King, for molecular genetics support. The genome scan was performed by the CIDR. The research was supported by National Institutes of Health grants NIDCD R01 DC03166, P30 DC04660, P01 DC00133, and NCRR M01 RR00400.
Appendix A: Criteria for Classification of Family Members as “Affected with COME/ROM”
Ear examination: Evidence of a positive history for COME/ROM was based on one or more of the following:
-
1.
retraction,
-
2.
perforation,
-
3.
atrophy,
-
4.
drainage,
-
5.
tympanosclerosis,
-
6.
tympanostomy tube,
-
7.
cholesteatoma, and
-
8.
monomer.
Tympanogram*: Family members were considered affected if there was presence of one or more the following:
-
1.
Age >10 years,
-
A.
Y1< 0.4 or Y1>1.7 mmho,
-
B.
TW2⩾180 decaPascals (daPa),
-
C.
resonant frequency <630 or >1,400 Hz, and
-
D.
abnormal van Huyse pattern at 630 or 1,400 Hz.
-
A.
-
2.
Age ⩽10 years,
-
A.
Y1<0.4 or Y1>1.0 mmho,
-
B.
TW2⩾160 daPa,
-
C.
resonant frequency <710 or >1,400 Hz, and
-
D.
abnormal van Huyse pattern at 710 or 1,400 Hz.
-
A.
Self-reported history of one or more of the following:
-
1.
history of tympanostomy tubes, three or more OM episodes in 1 year, or six episodes before age 6 years,
-
2.
tympanic membrane (TM) perforation,
-
3.
mastoidectomy,
-
4.
otorrhea,
-
5.
cholesteatoma,
-
6.
tympanosclerosisy, and
-
7.
tympanoplasty.
Medical record of presence of one or more of the following:
-
1.
tympanostomy tubes,
-
2.
otorrhea,
-
3.
OME >2 mo,
-
4.
TM perforation,
-
5.
cholesteatoma,
-
6.
tympanoplasty, and
-
7.
three or more episodes of OM in 1 year or six episodes before age 6 years.
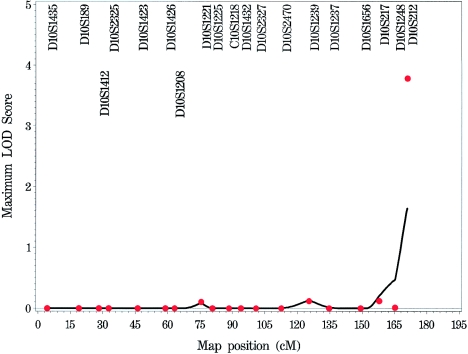
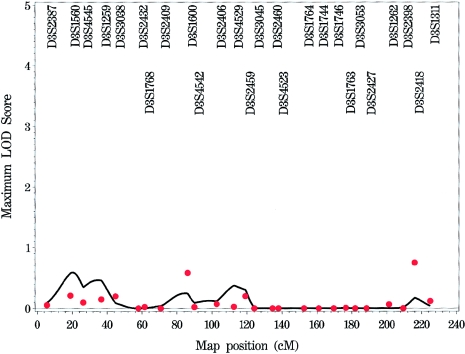
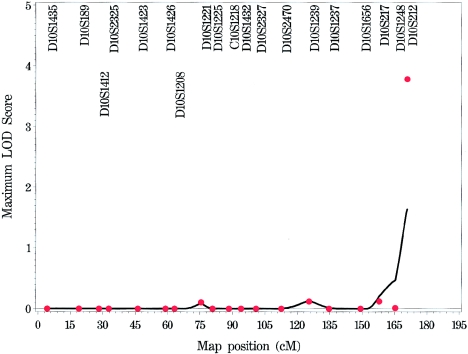
Appendix B: Genome Scan of COME/ROM for Each Chromosome
Figure A1.
Genome scan of COME/ROM for each chromosome, by use of multipoint (solid line) and single-point (stars) NPL models.
Footnotes
Y1 = static admittance. TW2 = tympanic membrane width.
Electronic-Database Information
The URL for data presented herein is as follows:
- University of California–Santa Cruz Genome Bioinformatics, http://genome.ucsc.edu/
References
- André P, Biassoni R, Colonna M, Cosman D, Lanier LL, Long EO, Lopez-Botet M, Moretta A, Moretta L, Parham P, Trowsdale J, Vivier E, Wagtmann N, Wilson MJ (2001) New nomenclature for MHC receptors. Nat Immunol 2:661 10.1038/90589 [DOI] [PubMed] [Google Scholar]
- Casselbrant ML, Mandel EM, Fall PA, Rockette HE, Kurs-Laskey M, Bluestone CD, Ferrell RE (1999) Heritability of otitis media: a twin/triplet study. JAMA 282:2125–2130 [DOI] [PubMed] [Google Scholar]
- Daly KA, Brown JE, Lindgren BR, Meland MH, Le CT, Giebink GS (1999) Epidemiology of otitis media onset by six months of age. Pediatrics 103:1158–1166 10.1542/peds.103.6.1158 [DOI] [PubMed] [Google Scholar]
- Daly KA, Rich SS, Levine S, Margolis RH, Le CT, Lindgren B, Giebink GS (1996) The family study of otitis media: design and risk factor profiles. Genet Epidemiol 13:451–468 [DOI] [PubMed] [Google Scholar]
- Ehrlich GD, Post JC (1999) Susceptibility to otitis media. JAMA 282:2167–2169 [DOI] [PubMed] [Google Scholar]
- Ho V, Daly KA, Hunter LL, Davey C (2002) Otoacoustic emissions and tympanometry screening among 0–5 year olds. Laryngoscope 112:513–519 10.1097/00005537-200203000-00020 [DOI] [PubMed] [Google Scholar]
- Hunter LL, Margolis RH, Rykken JR, Le CT, Daly KA, Giebink GS (1996) Extended high frequency hearing loss associated with otitis media. Ear Hear 17:1–11 10.1097/00003446-199602000-00001 [DOI] [PubMed] [Google Scholar]
- Janssens S, Beyaert R (2003) Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol Cell 11:293–302 10.1016/S1097-2765(03)00053-4 [DOI] [PubMed] [Google Scholar]
- Joki-Erkkilä VP, Puhakka H, Hurme M (2002) Cytokine gene polymorphism in recurrent acute otitis media. Arch Otolaryngol Head Neck Surg 128:17–20 [DOI] [PubMed] [Google Scholar]
- Jutel M, Watanabe T, Klunker S, Akdis M, Thomet OA, Malolepszy J, Zak-Nejmark, T, Koga R, Kobayashi T, Blaser K, Akdis CA (2001) Histamine regulates T-cell and antibody responses by differential expression of H1 and H2 receptors. Nature 413:420–425 10.1038/35096564 [DOI] [PubMed] [Google Scholar]
- King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, Witte DP, Rothenberg ME (2004) Expression and regulation of a disintegrin and metalloprotease (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol 31:251–265 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Kvaerner KJ, Tambs K, Harris JR, Magnus P (1997) Distribution and heritability of recurrent ear infections. Ann Otol Rhinol Laryngol 106:624–632 [DOI] [PubMed] [Google Scholar]
- Lahti M, Löfgren J, Marttila R, Renko M, Klaavuniemi T, Haataja R, Ramet M, Hallman M (2002) Surfactant protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr Res 51:696–699 10.1203/01.PDR.0000015911.65104.93 [DOI] [PubMed] [Google Scholar]
- Langefeld CD, Davis CC, Brown WM (2001) Nonparametric linkage regression I: combined European-American CSGA and German genome scans for asthma. Genet Epidemiol 21: S136–S141 [DOI] [PubMed] [Google Scholar]
- Löfgren J, Rämet M, Renko M, Marttila R, Hallman M (2002) Association between surfactant protein A gene locus and severe respiratory syncytial virus infection in infants. J Infect Dis 185:283–289 [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L (2000) Statistical tests for detection of misspecified relationships by use of genome-scan data. Am J Hum Genet 66:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rämet M, Löfgren J, Alho OP, Hallman M (2001) Surfactant protein-A gene locus associated with recurrent otitis media. J Pediatr 138:266–268 10.1067/mpd.2001.110133 [DOI] [PubMed] [Google Scholar]
- Rasmussen F (1993) Protracted secretory otitis media: the impact of familial factors and day-care center attendance. Int J Pediatr Otorhinolaryngol 26:29–37 10.1016/0165-5876(93)90193-7 [DOI] [PubMed] [Google Scholar]
- Rovers M, Haggard M, Gannon M, Koeppen-Schomerus G, Plomin R (2002) Heritability of symptom domains in otitis media: a longitudinal study of 1,373 twin pairs. Am J Epidemiol 155:958–964 10.1093/aje/155.10.958 [DOI] [PubMed] [Google Scholar]
- Roy S, Knox K, Segal S, Griffiths D, Moore CE, Welsh KI, Smarason A, Day NP, McPheat WL, Crook DW, Hill AV, Oxford Pneumococcal Surveillance Group (2002) MBL genotype and risk of invasive pneumococcal disease: a case control study. Lancet 359:1569–1573 10.1016/S0140-6736(02)08516-1 [DOI] [PubMed] [Google Scholar]
- Sipila M, Karma P, Pukander J, Timonen M, Kataja M (1988) The Bayesian approach to the evaluation of risk factors in acute and recurrent acute otitis media. Acta Otolaryngol 106:94–101 [DOI] [PubMed] [Google Scholar]
- Teele DW, Klein JO, Rosner B, the Greater Boston Otitis Media Study Group (1989) Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective cohort study. J Infect Dis 160:83–94 [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale, J (2000) Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA 97:4778–4783 10.1073/pnas.080588597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Higuchi Y, Yoshiyama K, Shimizu E, Kataoka M, Hijiya N, Matsuura K (1999) ADAM family proteins in the immune system. Immunol Today 20:278–284 10.1016/S0167-5699(99)01464-4 [DOI] [PubMed] [Google Scholar]
- Yamashita M, Fukui H, Sugama K, Horio Y, Ito S, Mizuguchi H, Wada H (1991) Expression cloning of a cDNA encoding the bovine histamine H1 receptor. Proc Natl Acad Sci USA 88:11515–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]