Abstract
By introducing beam-scanning multifocal multiphoton 4Pi-confocal microscopy, we have attained fast fluorescence imaging of live cells with axial super resolution. Rapid scanning of up to 64 pairs of interfering high-angle fields and subsequent confocal detection enabled us to perform three to five times finer optical sectioning than confocal microscopy. In conjunction with nonlinear image restoration, we demonstrate, to our knowledge for the first time, three-dimensional imaging of live eukaryotic cells at an equilateral resolution of ≈100 nm. This imaging mode allowed us to reveal the morphology and size of the green fluorescent protein-labeled mitochondrial compartment of live Saccharomyces cerevisiae (bakers' yeast) growing on different carbon sources. Our studies show that mitochondria of cells grown on medium containing glycerol as the only carbon source, as opposed to glucose-grown cells, exhibit a strongly branched tubular reticulum. We determine the average tubular diameter and find that it increases from 339 ± 5 nm to 360 ± 4 nm when changing from glucose to glycerol, that is, from a fermentable to a nonfermentable carbon source. Moreover, this change is associated with a 2.8-fold increase of the surface of the reticulum, resulting in an average increase in volume of the mitochondrial compartment by a factor of 3.0 ± 0.2.
Focused light is the only means by which it is possible to visualize and quantify biochemical processes in a live cell. Catalyzed by the advent of specific exogenous and endogenous markers, such as the green fluorescent protein (GFP) and its mutants, fluorescence-based far-field microscopy has evolved into a powerful tool for elucidating the relationship between cellular structure and function. This fact particularly applies to confocal and multiphoton fluorescence microscopes, which, by providing three-dimensional (3D) images, facilitate quantitative studies in the interior of cells (1). Despite its obvious advantages, the application of far-field microscopy has been seriously restricted. A major reason is the limited spatial resolution that has precluded 3D imaging and the quantitative 3D analysis of submicron-scale cellular compartments.
In Fourier space, where the imaging process is expressed in terms of spatial frequencies, the limited resolution of light microscopes is attributed to their inability to grasp and transfer object frequencies higher than about one-half of the reciprocal wavelength (2). In real space, the resolution limit is described by the minimal fluorescence spot size, which for a standard high-aperture confocal microscope is ≈180 and ≈500 nm in the transverse and axial directions, respectively (1). The demand for higher resolution in biological imaging has spurred a number of interesting developments, such as scanning 4Pi-confocal microscopy (3), stimulated emission depletion fluorescence microscopy (4, 5), wide-field I5M (6), and combinations of structured illumination with image restoration (7, 8).
In fluorescence imaging, the mathematical function describing the spot size is referred to as the point-spread function (PSF). Its Fourier transform is the optical transfer function (OTF) describing the spatial frequencies transferred by the microscope (2). Thus all concepts for improving spatial resolution aim at reducing the extent of the PSF in real space, which is equivalent to enlarging the extent of the OTF in Fourier space. Using the interference of the spherical wavefronts of two opposing lenses for illumination and/or confocal fluorescence detection, 4Pi-confocal microscopy renders a main maximum of the PSF three to seven times axially sharper and an OTF that is broader by the same factor (9–12). This concept is particularly powerful in connection with multiphoton excitation, because it substantially reduces undesired interference side lobes and provides a higher contrast of the interference pattern. Thus multiphoton 4Pi-confocal microscopy leads to a reliable axial resolution improvement (3, 13) of the order of 90–150 nm in both aqueously mounted (14) and glycerol-mounted fixed cells (13). Unfortunately, relying on stage scanning, whereby the sample is scanned slowly through the focus of the lenses, 4Pi-confocal microscopy has been hampered by slow data acquisition. Therefore, in the related I5M microscope (6), interferent illumination and detection are intentionally performed in the (nonconfocal) wide-field mode. In this instrument, data acquisition is substantially faster, so that the I5M is limited in speed only by a more extensive offline image restoration.
Comparative studies revealed, however, that the lack of confocal or multiphoton-induced suppression of the axial lobes is disadvantageous (12), because it results in high side lobes that are difficult to deal with through image processing (15). In the Fourier domain, the disadvantages are readily explained by the fact that the higher-frequency parts in the optical transfer function are only weakly connected to a strong central part. In addition, live cell imaging inevitably requires water-immersion lenses, which have a lower aperture angle (64.1°) than their oil counterparts (67.2°). As the performance of I5M and 4Pi-confocal microscopy seriously deteriorates with the aperture angle (12), the fragility of I5M most likely prohibits the use of water immersion with this system. As a result, axial superresolution in live cells and fast data acquisition have been difficult to combine (6). Although stimulated emission depletion microscopy has the potential to superresolve in 3D with a single lens (5), this concept requires the use of two pulsed-light trains of different wavelengths. Therefore, up to now, fast 3D light microscopy at ≈100-nm resolution has not been accomplished in live cell imaging.
In this work, we reconcile axial superresolution with fast live cell 3D imaging. For this purpose, we demonstrate parallelized multiphoton 4Pi-confocal microscopy, which allows up to 64 4Pi foci to be swiftly scanned across the specimen. This beam-scanning multifocal multiphoton 4Pi-confocal microscope, abbreviated here as MMM-4Pi, allowed us to cut down image acquisition time to seconds without compromising the super-resolving power. Equipped with an omnidirectional 100- to 140-nm resolution, we reveal the 3D morphology of GFP-labeled mitochondria in live yeast cells.
Multifocal Multiphoton 4Pi Microscopy and Resolution.
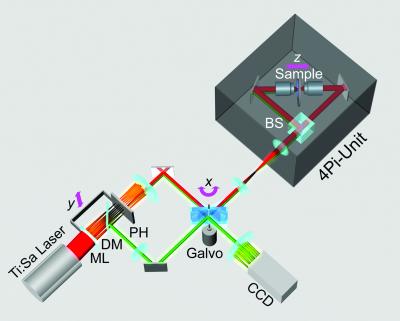
The sketch of the optical setup (Fig. 1) demonstrates that our MMM-4Pi microscope consists of two subunits. The head of the MMM-4Pi includes the beam splitter and the objective lenses, each illuminated through balanced interferometric arms. Axial scanning is performed by moving the sample along the optic axis (z), as is also the case with standard confocal microscopes. The multifocal beam-scanning arrangement is shown in the lower part of the drawing. The beam of a mode-locked Ti:Sapphire laser (Coherent, Palo Alto, CA) is split by an array of microlenses of focal length f = 10.7 mm, producing an n × m array of beamlets that, after passing through a dichroic mirror, are focused onto an array of pinholes. The pinholes act as point sources for creating the 4Pi illumination spots in the sample. After passing a pinhole, each beamlet is collimated by an achromatic lens and directed to a rapidly tilting galvanometric mirror. The following lenses ensure that the galvanometric mirror is imaged into the entrance pupils of the objective lenses. Rapid tilt of the mirror results in lateral scanning of the illumination 4Pi foci in the x direction.
Figure 1.
MMM-4Pi setup. An array of microlenses (ML) splits a pulsed laser beam into an array of beamlets, focused onto a pinhole (PH) array. After being cleaned by the pinholes, the beamlets are deflected by a scan mirror and directed to the 4Pi head, where an array of counterpropagating illumination foci is produced inside the specimen by using a beam splitter (BS). The array of fluorescence spots is imaged by the left objective lens back onto the pinhole array. The spatially filtered fluorescence is separated from the laser light by a dichroic mirror (DM) and scanned across the charge-coupled device camera by the back side of the mirror. Axial (z) scanning is accomplished by moving the specimen. Scanning in the y direction is performed by translating the linked pinhole and microlens arrays.
Scanning of the beamlets in the y direction is accomplished by translating the interlocked microlens and pinhole array. The fluorescence is collected by the left objective lens, deflected by the galvo mirror, and back-imaged onto the pinhole array; we refer to this imaging mode as 4Pi confocal of type A. The diameter of the pinholes corresponds to a 1.25 times back-projected Airy disk of the fluorescence light. This slight confocalization allows us to further suppress axial side lobes as well as residual aberrations stemming from the microlenses. After the filtering by the pinholes, the fluorescence light bounces off the long-pass dichroic mirror and is directed to the reflective back side of the galvo mirror, the tilt movement of which scans the array of fluorescence beamlets across a back-thinned charge-coupled device camera (MicroMax 512EBFT, Princeton Instruments, Trenton, NJ). Scanning the fluorescence beamlets with the back side of the galvo mirrors locks the scan of the excitation foci with those of fluorescence (16). n and m range between 4 and 8, so that a total of N = n × m = 16–64 parallel 4Pi foci is typical. Each of them performs ≈4 × 4-μm scans resulting in a field of view of ≈32 × 32 μm2.
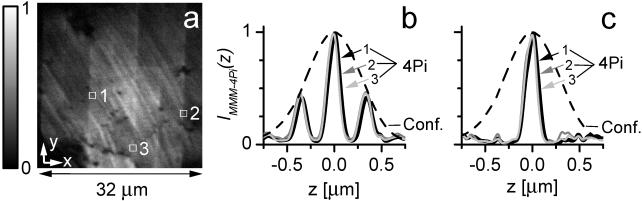
The parallelization is illustrated in Fig. 2a, showing a 32 × 32-μm2 xy image of a monomolecular fluorescent polydiacetylene layer; here we used an array of 8 × 8 foci. Whereas the thin diagonal stripes are due to the strongly polarized fluorescent backbone of the polydiacetylene, some of the visible vertical and horizontal lines mark the borders between individual scan fields. We emphasized these lines on purpose. They can be reduced by balancing the brightness of the foci and are apparent on uniform samples only; in most biological applications, they are largely negligible. The field size is readily changed by altering the tube lens or by changing the microlens and pinhole arrays. The number of parallel foci N depends on the available laser light. The camera is read out with a speed of typically one frame (256 × 256 pixels) per second.
Figure 2.
xy image of a fluorescent monolayer taken from a 3D stack (a). Axial intensity profiles through the 3D stack, IMMM-4Pi(z), exhibiting a sharp maximum and two lobes, recorded at three different coordinates (x,y) (b). The spatial invariance of the three profiles, IMMM-4Pi(z), indicates that the 4Pi-confocal PSF is constant over the whole field of view, irrespective of the microlens used. Fast linear one-step deconvolution can be applied throughout the field of view to remove the side lobes (c).
The speed of the MMM-4Pi depends on N, which in turn depends on the available laser power. The average power of 1–2 mW of a 4Pi spot leads to a focal peak intensity of ≈80 GW/cm2, which does not surpass the ≈200 GW/cm2 considered as an upper limit in nonlinear microscopy using ≈200-fs pulses (17). As a mode-locked Ti:Sapphire laser emits 1–2 W of average power, a parallelization by 1,000 foci rather than by n = 16–64 appears possible. At present, such a degree of parallelization is precluded by the losses connected with the cleaning of the laser and beamlet profile by the pinhole array.
The MMM-4Pi is functional only if the 4Pi PSF is invariant across the whole field of view. In fact, the possibility of fulfilling this condition has been called into question, because it requires joint movement of the 2N counterpropagating foci with a tolerance of less than 20 nm in the focal plane. In addition, the phase difference between the N pairs of interfering spherical wavefronts has to be identical during the scan process. We found that the field correction of modern water-immersion lenses fulfills this condition, as we demonstrate below. Hence, we can introduce an effective PSF of the MMM-4Pi-confocal microscope:
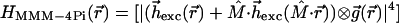 |
1 |
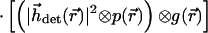 |
with
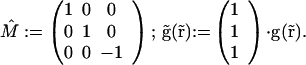 |
2 |
h⃗exc(r⃗) and M̂⋅hexc(M̂⋅r⃗) denote the counterpropagating focal fields generated by a single beamlet. M̂is a transformation matrix accounting for the counterpropagation. The resulting focal field is convoluted with the grid function g(x,y,0) of the array. The fourth power stems from the two-photon excitation of the fluorophore. The term |h⃗det(r⃗)|2 considers the imaging of the fluorescence onto the pinholes. The function p(r⃗) = p(x, y, 0) = 1 for 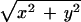 ≤ p0 takes into account their finite radius of p0.
≤ p0 takes into account their finite radius of p0.
A fundamental problem of any parallelized 3D microscope is the potential crosstalk between neighboring illumination and detection foci (18). If present, crosstalk deteriorates the axial resolution of the system by reinforcing specific planes above and beneath the focal plane. Crosstalk can be eliminated by time multiplexing, that is, ensuring that pulses of neighboring foci arrive at the sample at different time points (18–20). Calculating 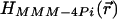 we found that in a MMM-4Pi with a quadratic grid arrangement, the 4Pi-foci can be brought as close as 3.5 λexc≈2.8 μm without leading to noteworthy crosstalk. This is because both two-photon excitation and confocal detection effectively suppress out-of-plane contributions. Hence, time multiplexing was not required to ascertain a spatial resolution similar to that of a stage-scanning single-beam 4Pi-confocal microscope. Parallelization does not compromise the resolution of our MMM-4Pi setup.
we found that in a MMM-4Pi with a quadratic grid arrangement, the 4Pi-foci can be brought as close as 3.5 λexc≈2.8 μm without leading to noteworthy crosstalk. This is because both two-photon excitation and confocal detection effectively suppress out-of-plane contributions. Hence, time multiplexing was not required to ascertain a spatial resolution similar to that of a stage-scanning single-beam 4Pi-confocal microscope. Parallelization does not compromise the resolution of our MMM-4Pi setup.
We found that a focal array facilitates the alignment of the two opposing lenses, because the necessity of matching two planes leads to well-defined alignment criteria. The MMM-4Pi in Fig. 1 relies on the interference of the illumination light only. Additional coherent superposition of the multiple fluorescence wavefronts at the charge-coupled device camera would be possible and would further improve axial resolution by another 30% (6, 9); however, the joint action of multiphoton excitation and slight confocalization already lead to a robust structure of the PSF with manageable lobe heights.
The structure of the PSF is readily confirmed in an experiment in which we quantify axial resolution through the z-response to an infinitely thin fluorescent layer: 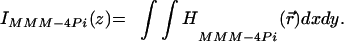 For this purpose, we recorded a stack of xy images of polydiacetylene layer at intervals Δz = 40 nm and integrated their intensity for each z coordinate. Fig. 2b depicts IMMM-4Pi(z) measured at three randomly chosen (x,y) coordinates in the image, produced by different beamlets. We find that our setup features a translational invariant 4Pi-confocal z-response with a pronounced main maximum and two lobes of ≈40%. The lobes are only 5% higher than in an optimized single-beam system (21). That they are well below 50% allows us to filter them out through a linear mathematical filter. The resulting effective z-response and axial resolution are shown in Fig. 2c, contrasted with the confocal version. The resolution improves from 700 ± 25 to 150 ± 10 nm in the MMM-4Pi. An important result is that the constructive interference is obtained throughout the field of view, proving clearly the viability of the parallelization of the 4Pi-confocal microscope in the fast beam-scanning mode.
For this purpose, we recorded a stack of xy images of polydiacetylene layer at intervals Δz = 40 nm and integrated their intensity for each z coordinate. Fig. 2b depicts IMMM-4Pi(z) measured at three randomly chosen (x,y) coordinates in the image, produced by different beamlets. We find that our setup features a translational invariant 4Pi-confocal z-response with a pronounced main maximum and two lobes of ≈40%. The lobes are only 5% higher than in an optimized single-beam system (21). That they are well below 50% allows us to filter them out through a linear mathematical filter. The resulting effective z-response and axial resolution are shown in Fig. 2c, contrasted with the confocal version. The resolution improves from 700 ± 25 to 150 ± 10 nm in the MMM-4Pi. An important result is that the constructive interference is obtained throughout the field of view, proving clearly the viability of the parallelization of the 4Pi-confocal microscope in the fast beam-scanning mode.
Imaging the Mitochondrial Network in Living Saccharomyces cerevisiae.
Mitochondria are essential organelles of eukaryotic cells. Their prominent function is to supply the cell with energy from oxidative phosphorylation. The yeast S. cerevisiae regulates the shape and size of the mitochondrial compartment depending on the carbon source. In yeast, this organelle consists of a branched tubular network, the continuity of which depends on a balanced frequency of fusion and fission events (22, 23). The strain BY4743 (24) used in this study exhibits relatively simple mitochondrial networks on fermentable carbon sources like glucose, whereas on nonfermentable sources such as glycerol, the network is branched and more elaborate. Although strain-dependent differences have been reported (25), this appears to be a typical behavior.
In the mid-1970s, extensive electron microscopy studies were performed to elucidate the plasticity of the mitochondrial network (26, 27). In the absence of a live cell imaging tool with high resolution, these studies inevitably relied on fixed sliced cells. Furthermore, these “tedious studies” (26) did not allow the creation of large numbers of 3D reconstructions. Since then, electron microscopy has dramatically progressed and provided invaluable insight into the inner structure of mitochondria (28). Still, it is not applicable to live cells.
By contrast, expression of matrix-targeted GFP allows visualization of living mitochondria through fluorescence (29). To demonstrate its continuous reticular structure, Nunnari et al. (30) have gained 3D images of the mitochondrial compartment by means of epifluorescence microscopy combined with image restoration. This method, as well as confocal fluorescence microscopy (25), allowed a distinction of glycerol- and glucose-grown yeast cell populations. However, in both cases, the limited spatial resolution rendered an equilateral 3D representation of the mitochondrial compartment impossible.
Our MMM-4Pi study on yeast mitochondria was performed with unsynchronized cultures of diploid cells (BY4743) grown to the midlogarithmic phase in batch cultures in either YPGlucose (1% yeast extract, 2% peptone, 2% glucose) or YPGlycerol (1% yeast extract, 2% peptone, 3% glycerol) full medium. The cells had been transformed with the plasmid pVT100U-mtGFP for constitutive expression of mitochondria-targeted GFP (29). For imaging purposes, after a washing step, the cells were embedded in PBS (137 mM NaCl/2.7 mM KCl/4.3 mM Na2HPO4/1.4 mM KH2PO4, pH 7.3) with 1% agarose and mounted between two coverslips. The 1% agarose was added to avoid spatial movement of the cells and also to slightly raise the refractive index of the aqueous medium from 1.334 to 1.341.
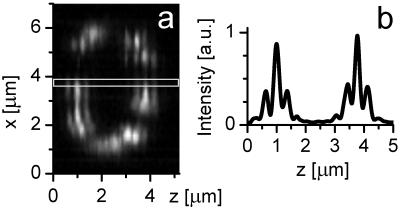
In Fig. 3, we exemplify the applicability of our microscope by displaying typical xz-image raw data; the excitation was performed at λexc = 890 nm. The data exhibit the effect of the sharp main peak and the 40% sidelobes (Fig. 3b). Similarly to Fig. 2b, nearly constructive interference is maintained throughout the image. Slight linear phase shifts induced by a residual mismatch in the refractive index between immersion liquid and medium were actively compensated (31). Occasionally, we observed slight changes of the relative phase because of local changes in the refractive index. However, these changes are rarely larger than π/6, at worst leading to slight shifts in the position of the tubules. Phase artifacts leading to noteworthy misrepresentations of the objects can be ruled out. Nonlinear image restoration further increases the resolution by 50% (13), so that we applied nonlinear image processing to all 3D sets shown further in the paper. The final transverse resolution of ≈140 and ≈100 nm along the optic axis is not yet sufficient to discriminate single cristae; however, the 3D representation of the tubular structure of the mitochondria is fundamentally improved.
Figure 3.
Raw data of an xz image (a) through a yeast cell with GFP targeted to the mitochondrial matrix and a representative intensity profile along the optic axis (b).
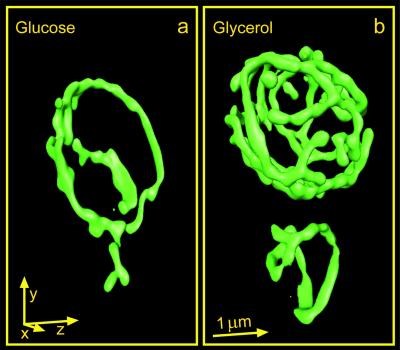
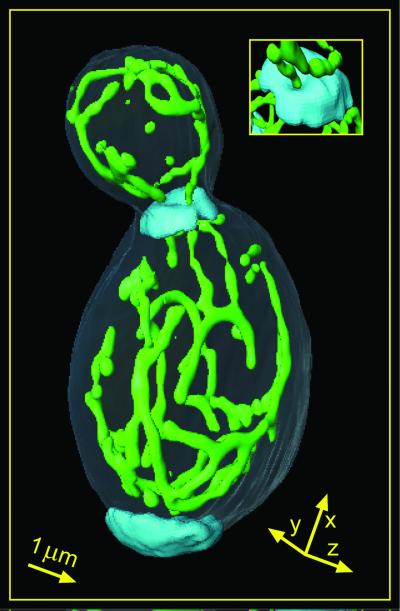
Fig. 4 a and b show typical surface-rendered reconstructions of 3D data sets of glucose- and glycerol-grown live cells, respectively. Comparison in Fig. 4 a and b reveals that the cells grown on glucose exhibit simple mitochondrial networks with few branch points. By contrast, the network of glycerol-grown cells is much more branched. These results are similar to those obtained with fixed cells by electron microscopy (26).
Figure 4.
GFP-labeled mitochondrial compartment of live S. cerevisiae (strain BY4743) recorded with the MMM-4Pi and displayed as surface-rendered 3D data. a and b show cells grown in complete medium with glucose or glycerol as the sole carbon source, respectively. An arrow corresponds to 1 μm in the respective direction.
Analysis of the Mitochondrial Network.
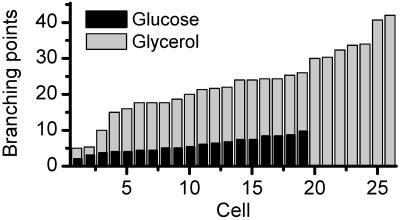
This difference is observed in the vast majority of cells in the respective populations. For a systematic analysis, we produced 19 and 26 data stacks of YPGlucose- and YPGlycerol-grown cells, respectively. Within a single cell, we always found a single ramified large mitochondrium and none or few smaller mitochondria. The 3D images enabled us to count the number of branch points of the mitochondrial network in each cell. They result from fusion between the tip of a mitochondrial tubule and a side of another one. In blind experiments, three individuals counted these branch points, obtaining virtually the same numbers. Fig. 5 displays the number of branch points for the glycerol- and glucose-grown cells, respectively. Whereas the glucose-grown cells featured 5.8 ± 2.1 branch points, we found 23 ± 9.3 in their glycerol counterparts, reflecting a mitochondrium that was four times more ramified. With three exceptions, none of the glycerol-grown cells featured less than 15 branch points, making their number a reliable hallmark of the two populations.
Figure 5.
Number of branch points within the mitochondrial network for the glucose- (black) and glycerol- (gray) grown cells, featuring 5.8 ± 2.1 and 23.0 ± 9.3 branch points, respectively.
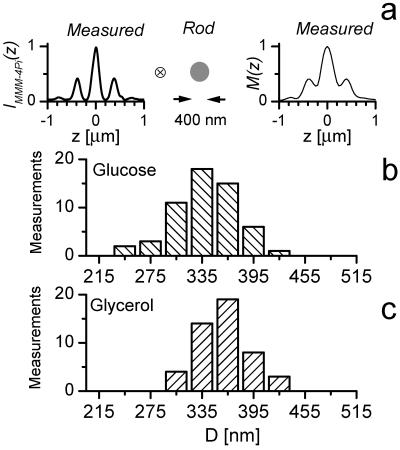
Matrix-targeted GFP has been shown to freely diffuse within the mitochondria (30, 32). Therefore, we can regard the tubules as flexible rods with uniform GFP distribution. Cristae invaginating into the matrix do not invalidate this assumption, because the volume of the cristae is small in comparison to that of the matrix. On the basis of these minimal premises, our microscope now allows us to precisely measure the thickness of individual tubules; this measurement is illustrated in Fig. 6a. Although in the MMM-4Pi a monomolecular layer produces a strongly modulated z-response IMMM-4Pi(z), a rod with diameter D produces a less modulated axial intensity profile M(z). Mathematically, M(z) is given by the convolution of IMMM-4Pi(z) with a function describing the rod. Clearly, for D > 150 nm, the lobe structure of M(z) will be blurred, and for D > 550 nm, it will even disappear. Fig. 6a exemplifies the measured IMMM-4Pi(z) along with a measured profile M(z) typically found in our data. The diameter D is now readily established by deconvolving M(z) with IMMM-4Pi(z). In fact, the evaluation can be simplified, because numerical simulation reveals that D can be directly concluded from the ratio between the average height of the side minima and side maxima. Under the conditions used, the diameter of single tubules was found to be 339 ± 35 nm and 360 ± 28 nm in the case of glucose- and glycerol-grown cells, respectively (Fig. 6 b and c). The increase in diameter per individual tubule is not significant; however, a large number of measurements renders the mean values of the two populations distinct. We found a 5.8 ± 1.7% increase in average diameter in the glycerol case (Fig. 6 b and c).
Figure 6.
Axial intensity profile through a mitochondrion M(z) exhibits elevated minima as a result of their finite diameter D. M(z) is represented by the convolution of a thin-layer response IMMM-4Pi(z) with a rod of diameter D, as sketched in a. Note that both M(z) and IMMM-4Pi(z) are measured data derived from a 3D image. Deconvolution analysis of the glucose- (b) and glycerol- (c) grown yeast cells reveals that in b, the average diameter is 339 ± 5 nm, whereas in c it is 360 ± 4 nm.
Sample-induced aberrations influence the modulation depth of the interference pattern of the excitation intensity. If the effect of aberrations were predominant over that of the convolution with the object function, the extraction of D would be impossible. Fortunately, the quadratic dependence of the fluorescence on the intensity counteracts the effect of aberrations and leads to deep, broad, and stable minima in IMMM-4Pi(z), as can be recognized from the left-hand profile of Fig. 6a. Therefore, in the MMM-4Pi, elevations of the side minima result nearly exclusively from finite tubular diameters. The measured profile M(z) in the right-hand panel of Fig. 6a is a good example, featuring both elevated side minima and broader peaks as compared with IMMM-4Pi(z). Modeling showed that this insensitivity against aberrations is hard to achieve in a single-photon excitation-based (interference) microscope. Therefore, the determination of spatial extents reported herein is a unique ability of the multiphoton excitation 4Pi-confocal approach.
As a next step, we investigated the change in the mitochondrial surface. Because the diffusing GFP renders a spatially uniform signal within the tubular rod, when scanning over a single tubule, fluorescence rises from a low value (the tubule is outside the focus) through a high signal (the tubule is filling the focus). Hence after normalization, the number of data voxels with a medium fluorescence intensity is proportional to the surface of the network. On the basis of this reasoning, the mitochondrial compartment of glycerol-grown cells features on average a 2.8 ± 0.2 times larger surface.
Combining this result with the diameter measurement, we find that the mean volume of the mitochondrial compartment of glycerol-grown cells is larger by a factor of 3.0 ± 0.2. An increase in volume means that additional lipid and protein biosynthesis is required when cells are transferred from a fermentable to a nonfermentable carbon source.
To relate the increase in mitochondrial volume to that of the cell, we also investigated the volume of the cells. We found that glycerol-grown cells are on average 1.5 times larger than their glucose-grown counterparts, which accounts for some of the increase in mitochondrial volume. Nevertheless, in the glycerol case, a larger proportion of the cell volume is occupied by the mitochondrial compartment, which is in good agreement with electron microscopy data (of a ≈10 times smaller set) of fixed cells (26).
The shape of the mitochondrial network, has been shown to change in the course of the cell cycle (33). However, these alterations are subtle compared with the differences induced by a change in carbon source. Hence, we decided to neglect them. To determine how the mitochondrial reticulum covers the cell periphery for each cell, we regarded the cell as a sphere consisting of 20 identical segments and calculated the overlap in volume of each of these segments with the GFP-labeled mitochondria. We found that the root mean square deviation from a uniform distribution is ≈0.48 and ≈0.86 for the glycerol- and glucose-grown cells, respectively (Table 1), indicating that, in the case of glycerol, the mitochondria are more evenly distributed in the cell.
Table 1.
Mean-square deviation of the fluorescence distribution (σ) is a measure of the inhomogeneity of the mitochondrial distribution. Featuring a higher σ, the mitochondrial compartments of glucose-grown S. cerevisiae are less uniformly distributed than their glycerol-grown counterparts
| Growth medium | σ |
|---|---|
| YPGlucose | 0.86 ± 0.20 |
| YPGlycerol | 0.48 ± 0.08 |
Mitochondria are known to be dynamic structures undergoing fusion and fission. Although the mitochondrial movement was potentially slowed down by mounting the cells, it was not abolished. As a typical stack of 65 xy images required 150 s, we cannot fully exclude mitochondrial translocation within the cell. However, as each xy image extends over the whole mitochondrial compartment, the acquisition time for a cellular xy section is rather short: 1-s exposure time plus 1.25-s camera readout time. Therefore, putative movements would have affected the z direction only. The average power per beamlet of 1–2 mW is associated with focal intensities of 40–80 GW/cm2, which is a comparatively low multiphoton excitation intensity. We routinely recorded more than three stacks of the same cell and never observed imaging-induced fragmentation of mitochondria, which is a typical sign of cellular distress.
To illustrate the localization of the mitochondrial network, in Fig. 7 we also imaged the cell wall and the bud scars. The cell wall was stained with calcofluor white (Sigma) and imaged in the standard two-photon mode at 800-nm excitation. Whereas the mitochondria were surface rendered, the intensely labeled bud scars were volume rendered. The sketch of the wall was performed by interactive contour tracing. The mother cell is prolate, as expected from a diploid strain. The cell features a large single highly ramified mitochondrium, which is exclusively located directly beneath the plasma membrane. No tubules cross the cell interior. It is interesting to note that the mitochondria are continuous from the mother cell to the bud. In fact, a mitochondrial tubule can be followed through a tube through the thickened cell wall at the budding site (see Fig. 7 Inset and Movie 1, which is published as supporting information on the PNAS web site, www.pnas.org). Evidently this cell is in an earlier stage of the cell cycle than the one shown in Fig. 4b, where the mitochondrial compartments between the mother and daughter cells are separated.
Figure 7.
S. cerevisiae with GFP-targeted mitochondrial matrix, grown on YPGlycerol. Displayed is a surface-rendered 3D-data stack of the mitochondrial network recorded with the MMM-4Pi. The cell wall and bud scars have been separately stained with the dye calcofluor white. The bud scars are volume rendered, whereas the visualization of the cell wall was performed by interactive contour tracing. The length of each arrow corresponds to 1 μm. (Inset) Magnification of the budding site from a different perspective. A mitochondrial tubule crosses the budding site through a channel in the bud scar (see Movie 1).
A fraction of the mitochondrial network accumulates in the bud tip, which probably reflects an attachment site, where the mitochondria are supposed to be immobilized on actin patches (33). Apparently the cell is budding for a second time, because another bud scar is found at the distal pole. This bipolar-budding pattern is typical for a/α cells (34).
Conclusion and Outlook.
Introducing MMM-4Pi, we have synergistically combined fast 3D imaging with axial superresolution. By deflecting an array of 4Pi foci across the specimen with a tilting mirror, we cut down the image acquisition time of 4Pi-confocal microscopy 10- to 15-fold. Conversely, our system can be regarded as the first successful improvement, to our knowledge, of the axial resolution of fast beam-scanning confocal microscopy.
The unique combination of speed, resolution, and live cell compatibility allowed us to access the 3D structure of the mitochondrial compartment in yeast cells with a 3D resolution in the 100-nm range, which is unprecedented in live eukaryotic cell imaging. The structure of the mitochondrial reticulum in living S. cerevisiae (strain BY4743) depends on the carbon source. In the case of glycerol-based medium, mitochondria display a strong ramification that is absent for mitochondria grown on a glucose medium. Live-cell compatibility of our microscope allowed us to measure the diameter of the tubules in the mitochondrial reticulum and to determine a 3-fold increase in volume because of the carbon source change. The precision of the thickness measurements is rooted in the nonlinear excitation mode in the MMM-4Pi, which avoids aberration-induced artifacts. We expect this method to be applicable to many other organelles of 150- to 550-nm spatial extent in live cells.
Further refining of the optical elements, as well as use of improved microlenses, will allow us to raise the number of foci to n = 100 and higher. In the MMM-4Pi, parallelization is facilitated by the doubling of the local intensity through interference of the focal fields. Moreover, recent developments in charge-coupled device detection, such as on-chip signal postamplification, will eliminate the finite readout time and enhance the sensitivity of our system by another factor of 5. Hence, we can anticipate that the imaging speed of our microscope will be further augmented by a factor of 20 in the near future, without any conceptual changes. As the current speed limitations are intrinsic neither to the 4Pi technique nor to the scanning system, we anticipate that our approach will open up superresolution 3D microscopy of live cells with data acquisition at video rate, which in turn is likely to play an important role in the elucidation of the relationship between structure and function in live cells.
Supplementary Material
Acknowledgments
We thank Dr. Benedikt Westermann (University of Munich) for helpful discussions and for the mitochondrial GFP-expressing BY4743 strain. Rainer Pick helped with the technical design of the MMM-4Pi setup. Furthermore, we thank Andreas Schönle for valuable discussions and for providing us with his image analysis software imspector 4d. We thank Nadia Martini and Jörg Bewersdorf for counting the branch points. This work was supported by the Deutsche Forschungsgemeinschaft through Grant He-1977.
Abbreviations
- GFP
green fluorescent protein
- 3D
three dimensional
- PSF
point-spread function
- MMM-4Pi
multifocal multiphoton 4Pi-confocal microscope
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pawley J. Handbook of Biological Confocal Microscopy. New York: Plenum; 1995. [Google Scholar]
- 2.Goodman J W. Introduction to Fourier Optics. New York: Mc Graw–Hill; 1968. [Google Scholar]
- 3.Hell S W, Stelzer E H K. Opt Commun. 1992;93:277–282. [Google Scholar]
- 4.Hell S W, Wichmann J. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 5.Klar T A, Jakobs S, Dyba M, Egner A, Hell S W. Proc Natl Acad Sc USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gustafsson M G L, Agard D A, Sedat J W. J Microsc. 1999;195:10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- 7.Gustafsson M G L. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- 8.Frohn J T, Knapp H F, Stemmer A. Proc Natl Acad Sci USA. 2000;97:7232–7236. doi: 10.1073/pnas.130181797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hell S, Stelzer E H K. J Opt Soc Am A. 1992;9:2159–2166. [Google Scholar]
- 10.Gu M, Sheppard C J R. J Opt Soc Am A. 1994;11:1619–1627. [Google Scholar]
- 11.Schrader M, Kozubek M, Hell S W, Wilson T. Opt Lett. 1997;22:436–438. doi: 10.1364/ol.22.000436. [DOI] [PubMed] [Google Scholar]
- 12.Nagorni M, Hell S W. J Opt Soc Am A. 2001;18:36–48. doi: 10.1364/josaa.18.000036. [DOI] [PubMed] [Google Scholar]
- 13.Hell S W, Schrader M, van der Voort H T M. J Microsc. 1997;185:1–5. doi: 10.1046/j.1365-2818.1997.2410797.x. [DOI] [PubMed] [Google Scholar]
- 14.Bahlmann K, Jakobs S, Hell S W. Ultramicroscopy. 2001;87:155–164. doi: 10.1016/s0304-3991(00)00092-9. [DOI] [PubMed] [Google Scholar]
- 15.Nagorni M, Hell S W. J Opt Soc Am A. 2001;18:49–54. doi: 10.1364/josaa.18.000049. [DOI] [PubMed] [Google Scholar]
- 16.Brakenhoff G J, Visscher K. J Microsc. 1992;165:139–146. [Google Scholar]
- 17.König K, Becker T W, Fischer P, Riemann I, Halbhuber K J. Opt Lett. 1999;24:113–115. doi: 10.1364/ol.24.000113. [DOI] [PubMed] [Google Scholar]
- 18.Egner A, Hell S W. J Opt Soc Am A. 2000;17:1192–1201. doi: 10.1364/josaa.17.001192. [DOI] [PubMed] [Google Scholar]
- 19.Buist A H, Müller M, Squier J, Brakenhoff G J. J Microsc. 1998;192:217–226. [Google Scholar]
- 20.Andresen V, Egner A, Hell S W. Opt Lett. 2001;26:75–77. doi: 10.1364/ol.26.000075. [DOI] [PubMed] [Google Scholar]
- 21.Schrader M, Hofmann U G, Hell S W. J Microsc. 1998;191:135–140. doi: 10.1046/j.1365-2818.1998.00361.x. [DOI] [PubMed] [Google Scholar]
- 22.Hermann G J, Shaw J M. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- 23.Yaffe M P. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]
- 24.Brachmann C B, Davies A, Cost G J, Caputo E, Li J C, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Visser W, van Spronsen E A, Nanninga N, Pronk J T, Gijs Kuenen J, van Dijken J P. A van Leeuw. 1995;67:243–253. doi: 10.1007/BF00873688. [DOI] [PubMed] [Google Scholar]
- 26.Stevens B J. Biol Cell. 1977;28:37–56. [Google Scholar]
- 27.Hoffmann H P, Avers C J. Science. 1973;181:749–751. doi: 10.1126/science.181.4101.749. [DOI] [PubMed] [Google Scholar]
- 28.Frey T G, Mannella C A. Trends Biochem Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 29.Westermann B, Neupert W. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Nunnari J, Marshall W F, Straight A, Murray A, Sedat J W, Walter P. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egner A, Schrader M, Hell S W. Opt Commun. 1998;153:211–217. [Google Scholar]
- 32.Partikian A, Olveczky B, Swaminathan R, Li Y X, Verkman A S. J Cell Biol. 1998;140:821–829. doi: 10.1083/jcb.140.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon V R, Karmon S L, Pon L A. Cell Motil Cytoskeleton. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Chant J, Pringle J R. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.