Abstract
A gal4-containing enhancer–trap called C309 was previously shown to cause subnormal courtship of Drosophila males toward females and courtship among males when driving a conditional disrupter of synaptic transmission (shiTS). We extended these manipulations to analyze all features of male-specific behavior, including courtship song, which was almost eliminated by driving shiTS at high temperature. In the context of singing defects and homosexual courtship affected by mutations in the fru gene, a tra-regulated component of the sex-determination hierarchy, we found a C309/traF combination also to induce high levels of courtship between pairs of males and “chaining” behavior in groups; however, these doubly transgenic males sang normally. Because production of male-specific FRUM protein is regulated by TRA, we hypothesized that a fru-derived transgene encoding the male (M) form of an Inhibitory RNA (fruMIR) would mimic the effects of traF; but C309/fruMIR males exhibited no courtship chaining, although they courted other males in single-pair tests. Double-labeling of neurons in which GFP was driven by C309 revealed that 10 of the 20 CNS clusters containing FRUM in wild-type males included coexpressing neurons. Histological analysis of the developing CNS could not rationalize the absence of traF or fruMIR effects on courtship song, because we found C309 to be coexpressed with FRUM within the same 10 neuronal clusters in pupae. Thus, we hypothesize that elimination of singing behavior by the C309/shiTS combination involves neurons acting downstream of FRUM cells
Keywords: reproductive behavior, C309 enhancer trap, shiTS transgene, traF transgene, inhibitory fru RNA transgene
Various portions of the CNS in Drosophila melanogaster are inferred to control separate elements of normal male courtship (e.g., refs. 1 and 2), in part by analysis of abnormal behavior (e.g., refs. 3–7). Some such studies have involved brain-behavioral analyses of the fruitless (fru) gene and its mutants (reviewed in ref. 8). Different fru mutants exhibit courtship subnormalities to varying degrees and at separate stages of the courtship sequence, depending on the mutant allele (e.g., refs. 9–12). Most fru mutants court other males substantially above levels normally exhibited by pairs or groups of wild-type males (e.g., refs. 12 and 13). The original fruitless mutation leads to spatially nonrandom decreases of fru-product presence (14, 15) within particular subsets of the normal CNS expression pattern (16, 17), which may be causally connected with the breakdown of recognition that is a salient effect of fru1 on male behavior (9, 12). fru-like courtship can be induced by the effects of a transgene that encodes GAL4 (a transcription factor derived from yeast). When this C309 enhancer trap was combined with a GAL4-drivable factor containing a dominant-negative, conditionally expressed variant of the shibire gene (shiTS), heat treatment of doubly transgenic males caused them to court females subnormally and to court other males vigorously (18). Although this strain had been termed a mushroom body enhancer trap in terms of the gal4 sequence it contains, being expressed “predominantly” within that dorsal-brain structure (19, 20), Kitamoto revealed that C309 drives marker expression in a widespread manner (18). Therefore, we sought to correlate various CNS regions in which this transgene is expressed with its effects on male behavior, emphasizing a search for “C309 neurons” that might overlap with elements of the FRUM pattern.
We also entertained the possibility that the C309/shiTS combination causes a mere caricature of fruitless-like behavior. Therefore, what would be the courtship effects of C309 driving a transgene that produces the female form of the transformer gene product? This TRA protein participates in posttranscriptional control of fru's primary “sex transcript,” so that FRUM protein is not produced in females (reviewed in ref. 8; also see refs. 16 and 21). If C309 and traF are naturally coexpressed in a subset of the to-be-analyzed neurons, feminization of the overlapping cells should eliminate this protein. We extended these transgenic experiments to target fruitless expression specifically by gal4 driving of an inhibitory RNA (IR) construct, which was generated with fru DNA by Manoli and Baker (22). Their experiments furnish one object lesson as to how “enhancer–trap mosaics” can delve into the neural substrates of a complex behavioral process, an approach commonly taken to manipulate brain structures and functions in courtship experiments (2–7). Because few genetic loci putatively identified by such transposons have been specified, the tactics we applied are in the context of CNS regions in which expression of a “real gene” is hypothesized to underlie well defined behaviors.
Materials and Methods
Supporting Information. For further details, see Tables 3–5 and Figs. 5 and 6, which are published as supporting information on the PNAS web site.
Stocks of D. melanogaster, Crosses, and Fly Handlings. Cultures were maintained as in ref. 23. Pure control males came from a Canton-S wild-type (WT) stock. Other control types were male progeny of a given transgenic strain (see below) crossed to Canton-S. Adult males and females were collected and stored as in refs. 12 and 23 (see below for exceptions). The enhancer–trap line C309 (19) is homozygous for a gal4-containing transposon inserted into chromosome 2; such females were crossed separately to males carrying the following transgenes: UAS-shiTS (homozygous on chromosome 3), which disrupts synaptic transmission in a heat-sensitive manner under the control of a given gal4-containing, neurally expressed transgene (24); UAS-traF (homozygous on chromosome 2), which, when GAL4-driven, causes the female form of transformer (tra) mRNA to be produced (e.g., refs. 3 and 4); UAS-fruMIR [inserted into both the second and third chromosomes, the former heterozygous for the transgene and In(2LR)O,Cy, the latter homozygous], designed to produce a double-stranded IR that blocks production of male (M)-specific protein encoded by the endogenous fru gene (22); and UAS-egfp (homozygous on chromosome 2), which encodes an “enhanced” nuclear form of GFP (25).
Most culture rearings occurred at 25°C; but those involving UAS-fruMIR were effected separately at 25°C and 29°C, because the hotter condition was reported to accentuate the inhibitory effects of this transgene (22). Histochemistry involving effects of traF or fruMIR on the presence of FRUM in C309-expressing neurons used females from a stock carrying both C309 and UAS-egfp on the second chromosome (generated by meiotic recombination), crossed to UAS-traF or to “double-insert” UAS-fruMIR males. Additional transgene combinations used females from a C309/C309/Cha-gal80/In(3LR)TM6B, Hu transgenic stock, crossed separately to UAS-shiTS, UAS-traF, UAS-fruMIR, or UAS-egfp males; triply transgenic progeny should have gal4 driving eliminated in neurons that coexpress gal80 (see ref. 26) under the control of regulatory sequences from the Cholineacetyltransferase (Cha) gene (see refs. 18 and 27).
Behavior. Basic courtship quantification. Audio/video recordings were obtained and processed as in refs. 12 and 23, but most of the current records were captured with a Sony VX2100 digital camera. For transgenic-male/WT-female pairings, the two types of flies were readily distinguishable despite the largely feminized external appearance of XY flies carrying C309/UAS-traF or C309/UAS-traF/Cha-gal80. For transgenic male/WT male observations involving UAS-shiTS or UAS-fruMIR, the two male types look the same, so each WT male had the tip of one wing clipped off at the time of collection. Males including UAS-shiTS were stored at 25°C (permissive temperature) before testing. For restrictive-temperature observations, a male- and food-containing tube was placed in a 30°C water bath for 20–40 min, then aspirated into a mating cell for recording at 30°C. For permissive-temperature controls, test males remained in food containers at 25°C before transfer into female-containing chambers at that temperature. Recordings were converted to computerized files, and behaviors were “logged” and analyzed by using lifesongx (http://lifesong.bio.brandeis.edu, compare ref. 28) to compute percentages of observation periods during which any interfly interactions occurred (courtship index, CI) or courtship wing displays (wing extension index, WEI).
Song sounds. Digitized audio tracks were logged then analyzed (as in refs. 12 and 23), leading to computations of the parameters specified in Table 3.
Mating behaviors. Attempted copulations, Mating-initiation latencies, and copulation successes were quantified for several fly pairs in a plastic device (see ref. 1), at 25°C or at 30°C for tests involving shiTS.
Courtship chaining. Eight to 10 males of a given genotype were grouped in a food vial upon collection, stored for 3–4 days (at 25°C or 20°C), and then hand-timer recorded at 25°C for the amount of time that at least three males spent courting one another during a 10-min observation period (as in refs. 12 and 13), leading to chaining index (ChI) values. A given male group involving shiTS (previously stored at 20°C) was observed at 25°C for ChI determination, then shifted to 30°C for 1–2 h, after which a subsequent ChI value was obtained at that temperature. General locomotion and flight performances. These were quantified as in ref. 12.
CNS Histology. The principal specimens were whole-mounted brains plus ventral nerve cords (VNCs) dissected from 4-day-old adult males (stored 15–20 per food vial). Additional studies involved developing Drosophila, as described in Fig. 6. The histological procedures were essentially those described in refs. 15, 16, and 29 (see Fig. 6 for details). The secondary antibody applied to recognize anti-FRUM primary (see ref. 16), was Cy5-conjugated anti-rat IgG. Neuronal marking was analyzed by using a Leica TCS SP2, scanning in the Cy5 channel for singly labeled brains (anti-FRUM) or scanning for Cy5 (red) and GFP (green) in colocalization assessments, which included conversion of red to magenta and merging the two channels to create white readout. For doubly labeled specimens, gal4/FRUM coexpression within a given neuronal cluster was quantified by projecting a Z series of 1- to 2-μm sections with both channels merged (see Fig. 6 for details). Coexpression values (see Table 2) were computed by taking the total numbers of white cells from the left and right side of the FRUM-cluster CNS region within a given transgenic specimen and dividing this value by the WT “adult-FRUM” neuronal count for that cluster (this denominator = 2× the relevant hemi-brain or hemi-VNC value from the leftmost data column in Table 2).
Table 2. FRUM-containing neuronal groups overlapping with expression of the C309 enhancer trap.
| Location | FRUM neuronal group, cluster number | Cluster name | Description of CNS region (numbers of adult FRUM neurons per hemi-brain or—VNC) | FRUM cells overlapping with C309, % (n) | traF-affected FRUM cells overlapping with C309, % (n) | fruMIR-affected FRUM cells overlapping with C309 at 25°C/29°C, % (n) |
|---|---|---|---|---|---|---|
| Brain | 2 | fru-aSP2 | Anterior portion of superior protocerebrum (50 ± 2, n = 7) | 25 (7) | 2 (6) | 15 (9)/10 (5) |
| 5 | fru-mAL | Medially located, just above antennal lobe (34 ± 1, n = 9) | 21 (7) | 2 (6) | 11 (9)/6 (8) | |
| 7 | fru-mcAL | Anterior to mechanosensory neuropile of antennal lobe, near esophagus (36 ± 1, n = 12) | 26 (9) | 2 (7) | 24 (8)/16 (8) | |
| 8 | fru-SG | Within subesophageal ganglion (10 ± 1, n = 6) | 23 (5) | 3 (7) | 24 (3)/18 (4) | |
| 13 | fru-pSP2 | Posterior portion of superior protocerebrum, near mushroom-body (MB) calyx (20 ± 1, n = 5) | 47 (5) | 7 (6) | 52 (5)/30 (5) | |
| 14 | fru-P | Broadly scattered in posterior brain, ventral to MB calyx (78 ± 3, n = 5) | 31 (5) | 3 (7) | 29 (5)/28 (4) | |
| VNC | 16 | fru-Pr | Prothoracic ganglion (29 ± 1, n = 7) | 14 (13) | 4 (6) | 11 (4)/9 (7) |
| 17 | fru-PrMs | Between pro- and mesothoracic ganglion (83 ± 2, n = 8) | 31 (9) | 4 (6) | 35 (7)/29 (6) | |
| 18 | fru-MsMt | Between meso- and metathoracic ganglion (37 ± 1, n = 6) | 30 (5) | 7 (4) | 28 (6)/28 (4) | |
| 20 | fru-Ab | Within abdominal ganglion (99 ± 5, n = 4) | 23 (5) | 3 (7) | 16 (8)/16 (4) |
The second and third columns list FRUM cluster numbers, each followed by the name of a given such neuronal group (compare ref. 16) within the brain or ventral nerve cord (VNC). The fourth column includes novel counts of adult neurons, determined by application of anti-FRUM to CNSs of wild-type (WT) males (average numbers of immunoreactive cells per hemi-CNS region, ± SEM; n, number of specimens scored). An additional series of C309/UAS-traF CNS specimens had anti-FRUM applied for single-labeling; numbers of immunoreactive neurons for the 10 clusters led to an average of 62%, comparing each traF-affected count to the corresponding WT male value (the smaller percentages yielding the 62% average ranged from 42 to 86, n values for these C309/UAS-traF specimens ranged from 6 to 9). Right three columns: percentages for C309/fru coexpression, computed as described in Materials and Methods. Control percentage overlaps (per CNS specimen, whose n values are in parentheses), fifth column. Experimental transgenic coexpression percentages: XY/C309/UAS-egfp/UAS-traF, sixth column; C309/UAS-egfp/UAS-fruMIR, seventh column, based on data collected separately from males reared at 25°C or 29°C. A one-way ANOVA on (on arcsine-transformed percentages) and subsequent pairwise comparisons revealed significant traF-induced overlap reductions (compared with C309/egfp values) for each cluster (P values < 0.05); for C309/fruMIR at 25°C, coexpression reductions were significant only for clusters 2, 5, and 20; at 29°C, fruMIR caused significantly lower coexpression for clusters 2, 5, 7, 13, and 16.
Statistics. Such analyses were performed on behavioral data and histological counts, the former arcsine or log-transformed unless the raw values defined a normal distribution (compare refs. 12 and 23). The data were subjected to ANOVAs and subsequent planned pairwise comparisons if appropriate; α levels were adjusted to correct for experiment-wise error (exemplified in ref. 12).
Results
Separate Components of Courtship Affected by Neurally Expressed Transgenes. Heat-treated Drosophila males in which UAS-shiTS expression was driven by the C309 transposon exhibited a significant decrease in courtship toward females compared with the low-temperature control (Fig. 1A). This finding confirms that of Kitamoto (18). Reducing the extent to which C309 drives shiTS spatial expression by adding the Cha-gal80 transgene to this genotype resulted in a minimal diminution of courtship directed at females (CI, 68 ± 3; n = 7) compared to WT or UAS-shiTS (30°C) values (Fig. 1 A). Spatial expression of the Cholineacetyltransferase (Cha) gene (determined by transgenic analysis) is reviewed in ref. 27. The relevance of this CNS pattern to sexual behavior was examined by Kitamoto (18), although he observed the triply transgenic type only in terms of Cha-gal80 effects on interactions among males in groups (compare ref. 13).
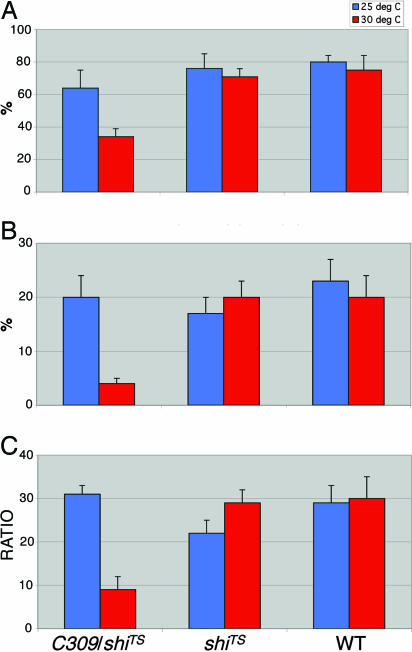
Fig. 1.
Male–female courtship affected by the C309 enhancer trap driving a temperature-sensitive shibire mutation. (A) CIs ± SEM for doubly transgenic C309/UAS-shiTS experimental males (C309/shiTS) and singly transgenic UAS-shiTS (shiTS) or wild-type (WT) control males, at the 2 temperatures indicated. C309 males paired with females at 25°C led to average CI and WEI values of 72 ± 3 and 34 ± 2, respectively (n = 8). n values for courtship values plotted: C309/shiTS, 9 (25°C) and 15 (30°C); shiTS, 10 and 15; WT, 14 and 9. A two-way ANOVA on CIs, with GENOTYPE and TEMPERATURE as the main effects, revealed significant differences for each effect (P values = 0.0003 and 0.037, respectively), but no interaction between these two factors (P = 0.15). (B) WEIs. A two-way ANOVA with GENOTYPE and TEMPERATURE as the main effects revealed significant WEI differences for each (P values = 0.0003 and 0.01, respectively) and a significant interaction effect between the two factors (P = 0.0005). (C) Ratios computed from the values in A and B.
These heat-induced courtship decrements are unlikely to be caused by overall behavioral sluggishness, because general locomotion scores at 30°C for C309/UAS-shiTS males (numbers of line crossings, ref. 12) were 64 ± 6 (n = 7), compared to 53 ± 5 (n = 10) for singly transgenic UAS-shiTS controls. Moreover, heated C309/UAS-shiTS males courted WT males at anomalously high levels (CI 16 ± 5, n = 15; vs. 4 ± 2 at 25°C, n = 8), belying the notion that 30°C leads to diminished vigor of this double-transgenic type. Typical CIs for WT male pairs are ≤4 (12–14). When the spatial expression of C309 was squeezed down by the effect of Cha-gal80, 30°C courtships performed by the triply transgenic males were still reasonably robust (CI, 10 ± 1; n = 11), compared, for example, with the behavior UAS-shiTS controls at 30°C (CI, 2 ± 0; n = 12) or C309/Cha-gal80 controls at 25°C (CI, 2 ± 1; n = 14).
Wing extensions measured for C309/UAS-shiTS males (Fig. 1B) plummeted relative to the overall courtship decrement (Fig. 1C), similar to the effects on WEI values of the fru3 or fru4 mutation (12). These fruitless mutants are not generally impaired in wing usage, because their flight performances were normal (12); however, heated C309/UAS-shiTS males flew subnormally. Whereas 44% of singly transgenic UAS-shiTS control males were adhered to the top 5 cm of the cylindrical test apparatus (12) and 25% were adhered to the bottom 30 cm, the corresponding values for C309/UAS-shiTS at 30°C males were 3% and 86%.
Courtship-song recordings of heated XY/C309/UAS-shiTS flies revealed them to generate almost no tone pulses; at 25°C, all males of this type sang vigorously with acoustical parameters in the normal range (Table 3). Taking into account the ≈9-fold reduction in wing extension for XY/C309/UAS-shiTS at 30°C relative to low-temperature controls (Fig. 1B) and the rate of song-pulse production for flies of this type at 25°C, one expects ≈18 pulses per minute if heat treatment would not impinge on singing. However, the 15 shiTS-affected males recorded at 30°C generated an average of only one pulse every 2 min (Table 3).
In context of nonmating being a key component of the fruitless syndrome (e.g., refs. 9 and 12), we found that only 17% of heated C309/UAS-shiTS males copulated; nearly all of them mated at permissive temperature (Table 4). Augmenting the current mating tests (18) showed that heated C309/UAS-shiTS males who copulated were not significantly different from controls in terms of mating-initiation latencies (Table 4), but quantification of attempted copulation revealed them to be substantially subnormal (Table 5). If C309/UAS-shiTS males performed this abdominal bending, it did not presage mating success, indicating that this transgenic type exhibits a monotonically stochastic decrease in the probability of proceeding from early, to middle, to late stages of the courtship sequence (also see Table 1).
Table 1. Courtship of transgenic males directed at females or other males.
| XY courters | Index | Female courtees | Male courtees |
|---|---|---|---|
| C309/UAS-traF | WT female (n = 18) | WT male (n = 15) | |
| CI | 76 ± 3 | 38 ± 7 | |
| WEI | 36 ± 3 | 4 ± 1 | |
| WT | WT female (n = 14) | C309/UAS-traF male (n = 15) | |
| CI | 80 ± 4 | 10 ± 3 | |
| WEI | 28 ± 4 | 1 ± 1 | |
| C309/UAS-fruMIR | WT female (n = 15) | WT male (n = 20) | |
| CI | 67 ± 3 | 27 ± 7 | |
| WEI | 31 ± 3 | 7 ± 3 | |
| WT | WT female (n = 14) | C309/UAS-fruMIR male (n = 20) | |
| CI | 80 ± 4 | 6 ± 1 | |
| WEI | 28 ± 4 | 0 ± 0 | |
| C309/UAS-traF/Cha-gal80 | WT female (n = 11) | WT male (n = 18) | |
| CI | 63 ± 5 | 17 ± 3 | |
| WEI | 31 ± 4 | 3 ± 1 | |
| WT | WT female (n = 14) | C309/UAS-traF/Cha-gal80 male (n = 18) | |
| CI | 80 ± 4 | 14 ± 3 | |
| WEI | 28 ± 4 | 1 ± 1 |
Males of the left-column genotypes had their courtships recorded in the presence of the target types indicated (courtees; WT, wild type). CIs and WEIs (compare Fig. 1) are quoted ± SEM. Additional control values from singly transgenic males: UAS-traF, CI: 80 ± 3, n = 10; UAS-fruMIR, CI: 75 ± 5, n = 10; Cha-gal80, CI: 64 ±6, n = 9. For male pairs, each individual's behavior was quantified separately. A one-way ANOVA on arcsine-transformed CIs from male behavior directed at females, with GENOTYPE as the main effect, revealed significant differences among genotypes (P < 0.0001). Subsequent comparisons showed that C309/UAS-traF and 25°C-reared C309/UAS-fruMIR (leading to the tabulated values) courted females at the same levels as did singly transgenic control males. C309/UAS-fruMIR reared at 29°C courted females to the same degree (CI: 48 ± 5, n = 14) as did singly transgenic UAS-fruMIR males (CI: 63 ± 4, n = 7) reared at 29°C. C309/UAS-traF/Cha-gal80 males courted females less vigorously than did C309/UAS-traF (P < 0.05) but indistinguishably from Cha-gal80. C309/UAS-fruMIR/Cha-gal80 males courted females (CI: 56 ± 4, n = 13) indistinguishably from the performance of C309/UAs-fruMIR (25°C) and the Cha-gal80 type but significantly less than did another singly transgenic control: UAS-fruMIR (P < 0.05). A one-way ANOVA on (untransformed) WEIs in the presence of females, with GENOTYPE as the main effect, revealed significant differences among genotypes (P = 0.008); but pairwise comparisons showed that most multiply transgenic types extended their wings to the same degree (P values ≥ 0.05) as did WT males or corresponding singly transgenic ones (WEI for UAS-traF: 29 ± 3, n = 10; for UAS-fruMIR reared at 25°: 37 ± 4, n = 10; same reared at 29°C: 25 ± 3, n = 7; Cha-gal80: 26 ± 4, n = 9); exception: C309/UAS-traF males gave significantly higher WEIs in the presence of females compared with WT male performance (P < 0.05). A one-way ANOVA on arcsine transformed CIs for behavior directed at males, with GENOTYPE as the main effect, revealed significant differences among genotypes (P < 0.0001). These comparisons factored in results from C309/UAS-fruMIR reared at 29°C (CI: 4 ± 1, WEI 0, n = 12). Subsequent pairwise comparisons showed that C309/UAS-traF males courted other males more vigorously than did C309/UAS-furMIR or C309/UAS-traF/Cha-gal80 (P values < 0.05). When paired with a male, the behavior of all multiply transgenic types (except for C309/UAS-fruMIR reared at 29°C) led to significantly higher CIs compared with singly transgenic controls (P values < 0.05). The latter male types did not court WT males vigorously (all CIs < 5 for: C309, n = 10; UAS-shiTS, n = 12; UAS-traF, n = 12; Cha-gal80, n = 11; UAS-fruMIR, n = 9 and 12, 25°C and 29°C, respectively). CIs derived from male-pair recordings involving C309/UAS-fruMIR or C309/UAS-traF/Cha-gal80 males were statistically equivalent to the behavior of C309/UAS-shiTS courting males at 30°C; whereas those recorded for C309/UAS-traF were significantly higher (P < 0.05). To compare courtship directed at WT males by the transgenic types to behavior of WT males directed at the transgenics, a final set of statistical comparisons (as above) revealed that most of the seven experimental types (the three multiple transgenics tabulated plus four additional ones noted above) courted WT males more vigorously than the latter courted the former (P values < 0.05); exceptions: C309/UAS-traF/Cha-gal80 and C309/UAS-fruMIR reared at 29°C, males of which courted WT males at the same level as the latter courted these two transgenic types.
The fruitless-like behaviors exhibited by C309/UAS-shiTS males could be a coincidence, prompting us to demasculinize C309-expressing neurons. We first combined C309 with UAS-traF, which encodes the TRA protein that blocks generation of FRUM in WT females (see Introduction). Despite C309 being advertised as a predominantly mushroom body (MB) enhancer trap (19, 20), it is notable that XY/C309/UAS-traF flies are largely transformed into external females (Fig. 5), one indication of C309 being expressed well beyond the MBs in the adult CNS (also see refs. 18 and 30).
Behavioral analysis of this transgene combination showed that XY versions of traF-affected flies courted females with no significant decrement (Table 1) and also sang normally (Table 3). However, in male-pair tests, XY/C309/UAS-traF flies courted WT males at fru-like levels (Table 1). These double-transgenic flies also elicited anomalously high levels of courtship from WT males (Table 1). Thus, might XY/C309/UAS-traF flies court other males in part because of self-stimulation? Owing to the fact that mutations at the fruitless locus do not cause courtship elicitation (12, 31), we disrupted FRUM production other than via manipulation of tra expression by combining C309 with UAS-fruMIR, surmising that this IR construct would do the job neurally but leave such flies in basically male state externally. These C309/UAS-fruMIR males exhibited marginal courtship decrements toward females (Table 1) but a significant reduction to 60% of the control value when C309/UAS-fruMIR males had been reared at 29°C (Table 1). However, there was no consequence to singing behavior of combining C309 with UAS-fruMIR (Table 3). The effects of fruMIR included a modest intrusion on mating performance after 25°C rearing and more substantial abnormalities for latency and copulation values after rearing this double-transgenic fly at 29°C (Table 4). Regarding intermale courtship, an individual XY fly in which fruMIR was putatively knocking down FRUM expression courted his WT male partner as vigorously as did the XY/C309/UAS-traF flies, but the effects of C309-driven fruMIR did not include anomalous elicitation (Table 1).
When the spatial extent of C309-encoded gal4 was again reduced by the effects of Cha-gal80, triply transgenic males carrying UAS-traF interacted with females normally but courted other males at an anomalously elevated level (Table 1). XY/C309/UAS-traF/Cha-gal80 flies are feminized externally (see Fig. 5) and elicited about the same level of WT-performed courtship as did the doubly transgenic XY/C309/UAS-traF type (Table 1). When Cha-gal80 was added to the C309/UAS-fruMIR combination, these males exhibited higher levels of courtship toward WT males than did singly transgenic controls; however, the former CI (11 ± 1, n = 18) was only about three times the corresponding “baseline” values (Cha-gal80, CI 4 ± 1, n = 11; UAS-fruMIR, 3 ± 1, n = 9), compared with the ≈8-fold enhancement caused by the C309/UAS-fruMIR combination (Table 1).
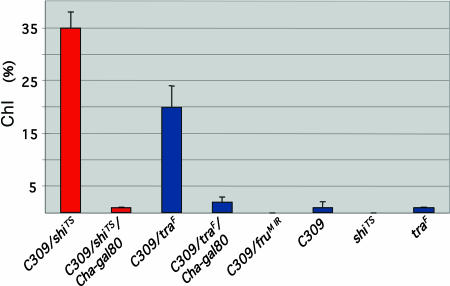
Courtship Among Males. The most striking fruitless-like behavior affected by the C309/UAS-shiTS combination is to induce interactions among several males grouped together (18). We also found that heated flies of this type form fru-like courtship chains (Fig. 2, compare refs. 12, 13, and 31). Males carrying C309 alone (but not UAS-shiTS by itself) gave a positive but tiny ChI (Fig. 2). This value was the same as that for C309/UAS-shiTS males to which the Cha-gal80 transgene was added (Fig. 2); the near elimination of C309/UAS-shiTS-induced chaining by Cha-gal80 further confirms the findings of Kitamoto (18).
Fig. 2.
Courtship chaining. For of C309/shiTS, 16 groups (8–10 males each) were observed; for C309/shiTS/Cha-gal80, n = 8. Grouped males were first measured at 25°C for ChI determinations (12, 13); these values were all 0. Fly containers were then shifted 30°C and had further ChIs determined. The C309/traF (n = 16) C309/UAS-fruMIR (n = 8) types and accompanying single-transgenic controls were always at 25°C. Additional observations of C309/UAS-fruMIR males were made after these flies had been reared at 29°C; the average ChI was 0 (n = 8 groups). One-way ANOVA on arcsine-transformed ChI values, with GENOTYPE as the main effect, revealed differences among groups (F[6,82] = 68.14, P < 0.0001). Subsequent pairwise comparisons showed that C309/shiTS at 30°C chained significantly more than C309/traF males (P < 0.05); both types exhibited much higher levels of chaining than the single-transgenic controls (P < 0.05). Although UAS-traF males (n = 10 groups) showed very brief moments of chaining, their behavior was not significantly different from that of UAS-shiTS (n = 17) or C309 (n = 11) control groups. The behavior of C309/fruMIR males was statistically equivalent to those exhibited by singly transgenic control males; the very low ChI values for groups of C309/traF/gal80 (n = 9) or C309/shiTS/gal80 males were not significantly different from the behavior of control males.
To ask whether the chaining behavior of C309/UAS-shiTS males can be apprehended in the context of sex-determination factors, this driver was combined with UAS-traF. Chaining was vigorous compared, for example, with the behavior measured for singly transgenic XY/UAS-traF groups (Fig. 2). Adding Cha-gal80 led to a ≈10-fold ChI decrement (Fig. 2) compared with the effect of unfettered C309 driving UAS-traF, even though C309 driving UAS-traF in the presence of Cha-gal80 induced courtship between males in single-pair tests (Table 1). Another transgene combination with that behavioral effect (Table 1) involves disruption of fru+ alone in C309/UAS-fruMIR males. However, groups of such flies exhibited no chaining behavior (Fig. 2).
CNS Expression of fruitless and a Transgene Marker. We found that C309 drove expression of GFP (Fig. 3) in nearly all CNS ganglia of adult males (see ref. 18). This gal4-containing transgene is expressed so broadly in central-brain and VNC regions that the pattern could encompass many CNS sites within which FRUM protein is normally found (see refs. 16, 17, and 32–34). We first determined the numbers of FRUM-immunoreactive neurons in WT adult males. These values were similar to the cell counts previously reported for pupal CNSs (16): for the 10 neuronal groups quantified (Table 2), the FRUM cell number was 6% greater (per cluster, on average) than the corresponding pupal values.
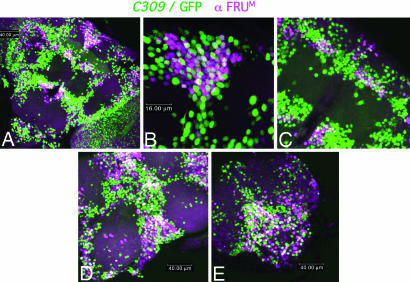
Fig. 3.
Coexpression of C309-driven marking and FRUM protein. Double labeling of CNS tissue dissected from C309/UAS-egfp male adults. Green denotes GFP-expressing cells; magenta denotes anti-FRUM-mediated staining; white denotes coexpressions of the two kinds of signals. These images resulted from scanning 1- to 2-μm sections, then projecting them into a Z-series. (A) Anterior view of adult male (magnification, ×46), showing white colocalization of C309- and fru-expressing cells in the brain (whose total width, between the two distal-most bilaterally symmetrical optic lobes, is ≈450 μm); cells within FRUM clusters 2, 5, 7, and 8 (see Table 2) are represented. (B) Neurons within cluster 2 at magnification ×3.9 that of A. (C) Clusters 5 and 7, magnification ×2 that of A. (D) Ventral nerve cord (VNC), magnification ×1.5 that of A: ventral view of the thoracic ganglia; pro-thoracic region is in the upper right; the mesothoracic region is in the center; part of the metathoracic region is near the bottom left. (E) Abdominal ganglion (posterior-most VNC region), magnification ×1.5 of that in A.
Double-labeling CNS preparations from C309/UAS-egfp males (Fig. 3) revealed the marker and fru expression patterns to overlap. Coexpression was observed in half of the relevant neuronal clusters (see refs. 16 and 32) and for subsets of the adult neurons within those 10 cell groups (Table 2). Vast numbers of “GFP-only” cells were observed in the immediate vicinity of most groups of doubly labeled neurons (as can be inferred by scrutinizing Fig. 3). We compared C309 and FRUM expression patterns during metamorphosis, shortly after fru's sex promoter is initially activated (16). Examination of CNS specimens dissected from C309/UAS-egfp pupae revealed overlap between GFP and FRUM signals in the same 10 clusters observed for adults, although the coexpressing patterns within pupae were not as extensive (Fig. 6).
To correlate anomalous intermale courtships observed for XY/C309/UAS-traF flies with the effects of this feminizing transgene on FRUM's presence in the CNS, we found that traF caused a dramatic decrement in, but not complete elimination of, immunoreactivity (Table 2, and compare Fig. 4 with Fig. 3). The effect of fruMIR on signals elicited by anti-FRUM in such C309 neurons was far less successful than traF, although high-temperature rearing increased the effect of inhibitory fru RNA (Table 2).
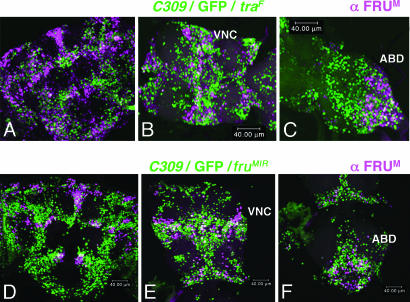
Fig. 4.
Diminished fruitless expression in the CNS caused by a feminizing or a demasculinizing transgene. Double labeling of CNS adult male tissue from C309/UAS-egfp/UAS-traF (A–C) and 25°C-reared C309/UAS-egfp/UAS-fruMIR (D–F). (A) Anterior view of traF-affected brain (magnification, ×40), showing marked reductions of coexpressing white cells within clusters 2, 5, 7, and 8, compared with the roughly corresponding images in Fig. 3; similar degrees of reduction appeared within: Pro-, meso-, and metathoracic VNC regions (B, displayed from left to right, ventral views at ×52 magnification), which contain FRUM clusters 16, 17, and 18 (Table 2). (C) Abdominal ganglion, containing VNC cluster 20 (at ×52 magnification). (D) Anterior view of fruMIR-affected brain at ×40 magnification; the reduction of coexpressing cells is not as dramatic as in A–C, as was the case for thoracic ganglia (E, ×40 magnification) and the abdominal ganglion (F, ×40 magnification).
When C309/UAS-egfp was combined with Cha-gal80, extents of C309-driven GFP overlapping with FRUM signals were diminished: instead of a 14–47% range for coexpressing cells (Table 2), the Cha-gal80-affected range was 4–37% (lowest value, cluster 7; highest value, cluster 13). This finding indicates that Cha's regulatory region (27) mediates CNS expression in large numbers of neurons in which C309's gal4 is active (also see ref. 18), but many of the latter cells would seem not to be cholinergic. The appreciable numbers of residual white cells not detectably affected by Cha-gal80 were scattered arbitrarily within the relevant FRUM clusters, compared with the possibility that the remaining doubly labeled neurons might have been bunched at discrete locations within a given neuronal group. Similarly, in C309/UAS-egfp/UAS-fruMIR males, the many coexpressing neurons (Fig. 4 D–F), which can be regarded as added to the much more limited pattern observed in the CNSs of C309/UAS-egfp/UAS-traF flies (Fig. 4 A–C), involved no cohesive intracluster localization of “extra” GFP-expressing cells in which this fru IR did not diminish FRUM signals.
Discussion
Mutations at the fruitless locus and the C309/UAS-shiTS transgene combination each cause similar courtship subnormalities and anomalies (Figs. 1 and 2 and Table 1). In this context, the C309 enhancer trap is expressed in many CNS neurons that contain male-specific FRUM protein (Fig. 3 and Table 2). One element of the courtship effects of C309/UAS-shiTS involves subnormal interactions between males and females. From correlating C309/FRUM coexpression with the fact that fruitless mutations lead to lower-than-normal male–female courtship (reviewed in ref. 35), we speculate that FRUM brain regions 2, 8, 13, and 14 (Table 2) are connected with the deleterious effects of shiTS. As to why two additional neuronal groups that coexpress FRUM and C309 are not noted here (groups 5 and 7), see below.
With respect to males courting females, special attention might be paid to clusters 13 and 14. These posteriorly located groups of FRUM neurons are likely to include brain regions within which genetic maleness is required if sex mosaics are to exhibit orientation toward and following of females (1, 2, 36). A problem with this interpretation is that feminizing substantial proportions of the C309/FRUM coexpressing cells led to no decrements in male–female interactions (Table 1), despite the 7- to 10-fold coexpression reductions caused by XY/C309/UAS-traF within clusters 13 and 14. Perhaps the relevant “overlap percentages” would have had to drop from 47 and 31 to 0 for both of these clusters (Table 2) if a traF-affected courtship decrement were to be realized. An alternative to viewing this matter in the context of C309/FRUM coexpression is that certain neurons in which this gal4 driver is active could be anatomically downstream of the fru-expressing brain cells that influence a male's ability to initiate and sustain courtship of a female.
This conception is relevant to the striking elimination of courtship song in recordings of C309/UAS-shiTS males. Once again, UAS-traF had no such effect (Table 3). Neurogenetic findings pertinent to this matter are that C309 is expressed in imaginal thoracic ganglia (Fig. 3 and Table 2); fru is “song-involved” (12, 37); this gene makes its products within several regions of the ventral nerve cord (16, 17, 32–34), coexpressing C309 within most of them (Table 2); and genetic maleness within Drosophila's VNC has been implicated in song control (reviewed in ref. 35). Thus, turning off synaptic transmission emanating from one or more subsets of the C309/UAS-shiTS neurons in the thoracic ganglia could be the etiology of heat-induced songlessness exhibited by these doubly transgenic males. Regarding the absence of a traF effect, what if C309 was not expressed in any FRUM-containing song-relevant neurons during metamorphosis? In other words, C309 expression in VNC neurons underlying song control could be activated late in the life cycle, allowing for the shiTS effect to take hold after adult males are heated; however, the progenitors of such cells might not express C309 during an earlier “feminization-relevant” stage, so that post-metamorphic activation of traF would occur too late to affect singing ability. However, we found substantial coexpression of FRUM and C309 within the pupal VNC (Fig. 6). In this respect, we submit that assessing the C309's expression pattern throughout the life cycle is a valuable object lesson as to what must be done properly to interpret the biological effects of a given enhancer trap.
As to the divergent effects C309-driven shiTS vs. traF, recall that the former factor seems broadly to impinge on VNC functioning, in that the fly's general ability to vibrate its wings is shut down by the synaptic disruptor; in contrast, songless fruitless mutants fly normally (12). Thus, consider a scenario in which C309 neurons would include those that mediate wing vibrations during flight, and that this transgene is expressed in separate VNC cells hypothetically dedicated to such vibrations during courtship. Therefore, we speculate that the expression domain of C309 includes inter- or motor-neurons functioning within and downstream of a “command center” for flight as well as neurons located in relatively distal regions of a separate anatomical pathway. The latter would originate where fru-expressing cells exert the gene's crucial regulation of courtship song.
Turning to anomalous courtship interactions among males, our focus shifts back to the brain: FRUM clusters 5 and 7, where fru1 causes an apparent absence of this protein; this mutation minimally affects the gene's expression in other brain regions (15). It is notable that the C309/UAS-traF combination knocked down driver/FRUM coexpression to ≈10% normal in cluster 5 (Table 2). Cluster 7 was similarly affected, but special attention should be paid to cluster 5 (see below). One reason that that group was surmised to be the etiology of frantic courtship among fru1 males (15) is that cluster 5 is located near the antennal lobes; and transgenically mediated feminization of a brain region near these structures induces intermale courtships (ref. 3, although none of the gal4 drivers in that study included C309). Therefore, if proper male-specific structure or function of cluster 5 is involved in normal sex recognition, the mutation's demasculinizing effect on this brain region, or transgene-effected feminization of it, could cause this aspect of courtship to break down.
Elements of the current findings (and those in ref. 18) suggest that abnormal formation of the brain region in question is not necessary for it to mediate anomalous interactions between males (also see refs. 33 and 34); this is because deactivating synaptic transmission in cluster 5 after CNS development completed itself in a male manner is sufficient to induce intermale courtship. Perhaps this behavioral effect of driving UAS-shiTS involves removal of inhibitory neurotransmission relevant to the functioning of this brain region (but see ref. 7), which in normal males would block their wherewithal to sustain courtship between males. Therefore, the fru1 effect on cluster 5 and that of driving TRA production in this region might not involve the formation of a sex recognition center (such that a hypothetical circuit involved in inhibiting intermale courtship is not present or miswired), but instead the intracellular quality and function of neurons in the mature brain.
Considering further that certain cluster 5 neurons comprise the subset of FRUM's spatial domain for shiTS- or traF-induced intermale courtship, the relevant cells would be those in which both fruitless and C309 generate their gene products (20% of the 35 neurons within this group: Table 2). One problem with this supposition is that C309/UAS-traF flies elicit fairly high levels of courtship (Table 1). Thus, groups of C309/UAS-traF males may form chains for reasons extending beyond a given fly's “motivation” inappropriately to court another male: The extent to which a C309/UAS-traF fly is feminized (Fig. 5) could include self-stimulation that might contribute to intermale courting. However, recall the case of C309/UAS-traF/Cha-gal80 males, a transgenic type that is similarly feminine externally and elicits courtship (Table 1). The diminished extent to which C309's gal4 is effective when combined with Cha-gal80 led to weakened homosexual courtship in single-pair tests (Table 1) and dramatically reduced chaining behavior (Fig. 2), although there was essentially no effect of Cha-gal80 on the basic courtship ability of these triply transgenic males. Thus, the effects of this “neurons-only” manipulation suggest that hypothetical self-stimulation, which did not cause C309/UAS-traF/Cha-gal80 males vigorously to court other ones, is minimally operating to induce the homosexual courtships performed by XY/C309/UAS-traF flies. Males carrying C309 and UAS-fruMIR are also not feminized externally; however, they courted other males robustly in single-pair tests, an effect that was diminished by adding Cha-gal80 (Table 1). Therefore, we surmise that flies carrying a given fruitless-affecting transgene (Table 2) exhibit intermale courtship because the relevant CNS neurons are demasculinized.
However, what about neural structures not analyzed in the current study that could be involved in the behavioral effects of C309 driving either traFor fruMIR? Thus, consider that TRA affects the primary transcript emanating from the doublesex (dsx) gene (reviewed in ref. 8) and that dsx null mutations cause XY flies to exhibit modest levels of intermale courtships (38). C309 driving of traF could lead to the female (F) form of DSX (thus, no DSXM, as in dsx–) within brain cells connected to sex recognition other than those we analyzed. Indeed, dsx+ is expressed in the brain (39); however, the functional significance of these cells is unknown, let alone whether any of them also express fru+. In this regard, it was important to home in on disruption of fruitless's CNS expression alone by combining C309 with the UAS-fruMIR transgenes; this was sufficient to induce courtship between a given pair of doubly transgenic males (Table 1) but led to no chaining (Fig. 2). Thus, anomalously high levels of courtship between two males has been disconnected from courtship chaining. [The same disconnect between these different kinds of intermale courtship occurred when Cha-gal80 was added to the C309/UAS-traF combination (Table 1 vs. Fig. 2).] It is as if the broad neural effects of a genetic abnormality such as a fruitless mutation, or combining C309 with UAS-traF, is necessary to cause sustained courtship among several variant males; however, if the impingement on fru+ expression is more limited (right column of Table 2), only courtship between a pair of males can occur.
In this regard, the C309/UAS-fruMIR flies were substantially less affected in terms of numbers of brain neurons within which FRUM became undetectable, compared with the effect of the same driver combined with UAS-traF (Table 2). This brings us to the matter of additional neurons that are potentially relevant to courtship and should be analyzed in context of the C309 effects. Here, we refer to the many PNS cells recently discovered to express fruitless in external sensory structures (33, 34). It is unknown whether any of these neurons coexpress C309, such that sensory inputs relevant to courtship may have been impinged upon by combining that transgene with UAS-shiTS or with the sex-affecting transgenes. However, fru+ expression in external appendages is not required for a fly to recognize, follow, and perform wing extension at a female: when these structures are genetically female in certain gynandromorphs, maleness within the brain is sufficient to trigger mosaic-with-female courtship (1, 36).
The relative inefficacy of one of the C309-driven transgenes just alluded to warrants further comment: The investigators who generated the fru IR construct reported that a gal4 driver, when combined with two doses of UAS-fruMIR, knocked out detectability FRUM throughout brain cluster 7, against a backdrop of all neurons within this group coexpressing fru+ within the adult male's brain (22). The C309 driver, which may be weaker than that applied by Manoli and Baker (22), leads to overlapping expression with FRUM in only a subset of cluster 7 neurons. The C309/UAS-traF combination dropped the relevant coexpression percentage 13-fold (Table 2), but such a decrement did not lead to inappropriately rapid courtships and merging of courtship steps: the behavioral anomalies reported by Manoli and Baker appear to be caused by knocking out FRUM in all cluster 7 neurons by combining their driver with either UAS-traF or UAS-fruMIR. Importantly, “cluster 7 FRUM-null” males did not court other males (22).
The current study aimed to delve into various regions of the male CNS in which the fruitless gene is expressed: Do certain subsets of the spatial pattern govern a male's ability to perform a discrete feature of the reproductive sequence? Using the gal4-containing C309 enhancer trap was valuable, because it leads to impersonations of certain fru-mutant behaviors when this driver is combined with a shiTS-containing factor that broadly disrupts neural functioning. By limiting C309's efficacy to disrupt by causing it to drive sex-related transgenes succeeded in provisionally partitioning fru-related “sex recognition” neurons to a subset of the normal brain pattern. By subtraction, the partitioning was further delimited by knocking out the driver's efficacy in a subset C309's spatial domain: adding a neurally driven gal80 transgene that substantially (Table 1) or completely (Fig. 2) attenuated anomalous intermale courtships. A pleasant surprise occurred when the C309/UAS-fruMIR combination was found not to mimic the effects on courtship among males of combining the driver with UAS-traF. Thus, the broader pattern of FRUM expression, unaffected by the IR compared with the substantial decrement caused by traF, takes the analysis a further step. For example, we have begun to tease out the manner by which fru mutations and related factors influence courtship between two males, as opposed to the much more complicated behavioral dynamics that can occur in a group of such Drosophila.
However, inferences about the potentially relevant subsets of a given brain cluster do not approach specifically identifiable neurons. For this, it will be necessary to do more than quantify the cells in which a transgene driver and fruitless are coexpressed. Further brain-behavioral dissections will require assessing the differential connectivity patterns defining a given class of FRUM neurons, along with variations of cellular content that are likely to discriminate one category of such neurons from another. The relevant object lessons stem from analyses of, so far, only the posterior-most component of fruitless's expression domain in the male CNS: partitioning certain abdominal-ganglion neurons that differentially connect with either a male-specific muscle (17, 32) or with internal reproductive organs (29, 32), and discovering that the latter type of FRUM cells uniquely contain serotonin (15, 29). Neurons containing another neurotransmitter, acetylcholine, are on point; but not all of the C309 effects can be ascribed to neurons affected by Cha-gal80, because (in contrast to ref. 18) we found certain courtship defects to remain when analyzing males that carried this transgene along with C309 and UAS-shiTS. This finding reinforces that notion that additional neuronal qualities must be uncovered with regard to cells expressing this enhancer–trap, the fruitless gene, or both.
Supplementary Material
Acknowledgments
We thank Devanand Manoli (Stanford University, Stanford, CA) for providing the fruMIR transgenic strain, John Reiffel for upgrading courtship-analysis software, Joseph Sacks for helping with behavioral analysis, Edward Dougherty for confocal assistance, and Jennifer Mehren for comments on the manuscript. This work was supported by National Institutes of Health Grants GM-21473 and NS-33352.
Author contributions: A.V. and J.C.H. designed research; A.V., S.L.F., and J.D.K. performed research; J.C.H. contributed new reagents/analytic tools; A.V., S.L.F., and J.D.K. analyzed data; and A.V. and J.C.H. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
Abbreviations: IR, inhibitory RNA; CI, courtship index; WEI, wing extension index; ChI, chaining index; VNC, ventral nerve cord.
See accompanying Profile on page 16547.
References
- 1.Hall, J. C. (1979) Genetics 92, 437–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferveur, J.-F. & Greenspan, R. J. (1998) J. Neurogenet. 12, 205–226. [DOI] [PubMed] [Google Scholar]
- 3.Ferveur, J.-F., Störtkuhl, K. F., Stocker, R. F. & Greenspan, R. J. (1995) Science 267, 902–905. [DOI] [PubMed] [Google Scholar]
- 4.O'Dell, K. M. C., Armstrong, J. D., Yang, M. Y. & Kaiser, K. (1995) Neuron 15, 55–61. [DOI] [PubMed] [Google Scholar]
- 5.Heimbeck G, Bugnon, V., Gendre, N., Keller, A. & Stocker, R. F. (2001) Proc. Natl. Acad. Sci. USA 98, 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kido, A. & Ito, K. (2002) J. Neurobiol. 52, 302–311. [DOI] [PubMed] [Google Scholar]
- 7.Broughton, S. J., Kitamoto, T. & Greenspan, R. J. (2004) Curr. Biol. 14, 538–547. [DOI] [PubMed] [Google Scholar]
- 8.Baker, B. S., Taylor, B. J. & Hall, J. C. (2001) Cell 105, 13–24. [DOI] [PubMed] [Google Scholar]
- 9.Hall, J. C. (1978) Behav. Genet. 8, 125–141. [DOI] [PubMed] [Google Scholar]
- 10.Ito, H., Fujitani, K., Usui, K., Shimizu-Nishikawa, K., Tanaka, S. & Yamamoto, D. (1996) Proc. Natl. Acad. Sci. USA 93, 9687–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryner, L. C., Goodwin, S. F., Castrillon, D. H., Anand, A., Villella, A., Baker, B. S., Hall, J. C., Taylor, B. J. & Wasserman, S. A. (1996) Cell 87, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 12.Villella, A., Gailey, D. A., Berwald, B., Ohshima, S., Barnes, P. T. & Hall, J. C. (1997) Genetics 147, 1107–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, G. & Hall, J. C. (2000) Behav. Genet. 30, 263–275. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin, S. F., Taylor, B. J., Villella, A., Foss, M. Ryner, L. C., Baker, B. S. & Hall, J. C. (2000) Genetics 154, 725–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, G. & Hall, J. C. (2001) J. Neurosci. 21, 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, G., Foss, M., Goodwin, S. F., Carlo, T., Taylor, B. J. & Hall, J. C. (2000) J. Neurobiol. 43, 404–426. [DOI] [PubMed] [Google Scholar]
- 17.Usui-Aoki, K., Ito, H., Ui-Tei, K., Takahashi, K., Lukacsovich, T., Awano, W., Nakata, H., Piao, Z. F., Nilsson, E. E., Tomida, J.-y. & Yamamoto, D. (2000) Nat. Cell Biol. 2, 500–506. [DOI] [PubMed] [Google Scholar]
- 18.Kitamoto, T. (2002) Proc. Natl. Acad. Sci. USA 99, 13232–13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly, J. B., Roberts, I. J., Armstrong, J. D., Kaiser, K., Forte, M., Tully, T. & O'Kane, C. J. (1996) Science 274, 2104–2107. [DOI] [PubMed] [Google Scholar]
- 20.Dubnau, J., Grady, L., Kitamoto, T. & Tully, T. (2001) Nature 411, 476–480. [DOI] [PubMed] [Google Scholar]
- 21.Demir, E. & Dickson, B. J. (2005) Cell 121, 785–794. [DOI] [PubMed] [Google Scholar]
- 22.Manoli, D. S. & Baker, B. S. (2004) Nature 430, 564–569. [DOI] [PubMed] [Google Scholar]
- 23.Chan, B., Villella, A., Funes, P. & Hall, J. C. (2002) Genetics 162, 135–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamoto, T. (2001) J. Neurobiol. 47, 81–92. [DOI] [PubMed] [Google Scholar]
- 25.Barolo, S., Carver, L. A. & Posakony, J. W. (2000) BioTechniques 29, 726–732. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. M. & Luo, L. Q. (2001) Trends Neurosci. 24, 251–254. [DOI] [PubMed] [Google Scholar]
- 27.Yasuyama, K. & Salvaterra, P. M. (1999) Micscrosop. Res. Tech. 45, 65–79. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein, A. S., Neumann, E. K. & Hall, J. C. (1992) J. Insect Behav. 5, 15–36. [Google Scholar]
- 29.Lee G., Villella, A., Taylor, B. J. & Hall, J. C. (2001) J. Neurobiol. 47, 121–149. [DOI] [PubMed] [Google Scholar]
- 30.Waddell, S., Armstrong, J. D., Kitamoto, T., Kaiser, K. & Quinn, W. G. (2000) Cell 103, 805–813. [DOI] [PubMed] [Google Scholar]
- 31.Gailey, D. A. & Hall, J. C. (1989) Genetics 121, 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Billeter, J. C. & Goodwin, S. F. (2004) J. Comp. Neurol. 475, 270–287. [DOI] [PubMed] [Google Scholar]
- 33.Manoli, D. S., Foss, M., Villella, A., Taylor, B. J., Hall, J. C. & Baker, B. S. (2005) Nature 436, 395–400. [DOI] [PubMed] [Google Scholar]
- 34.Stockinger, P., Kvitsiani, D., Rotkopf, Tirián, L. & Dickson, B. J. (2005) Cell 121, 795–807. [DOI] [PubMed] [Google Scholar]
- 35.Hall, J. C. (2002) J. Neurogenet. 16, 135–163. [DOI] [PubMed] [Google Scholar]
- 36.Hall, J. C. (1977) Behav. Genet. 7, 291–312. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler, D. A., Kulkarni, S. J., Gailey, D. A. & Hall, J. C. (1989) Behav. Genet. 19, 503–528. [DOI] [PubMed] [Google Scholar]
- 38.Villella, A. & Hall, J. C. (1996) Genetics 143, 331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, G., Hall, J. C. & Park, J. H. (2002) J. Neurogenet. 16, 229–248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.