Abstract
RNase P complexes have been proposed as a novel RNA-based gene interference strategy to inhibit gene expression in human malignancies and infectious diseases. This approach is based on the sequence-specific design of an external guide sequence (EGS) RNA molecule that can specifically hybridize to almost any complementary target mRNA and facilitate its cleavage by the RNase P enzyme component. We designed a truncated RNase P-associated EGS molecule to specifically recognize the U5 region of HIV-1 mRNA and mediate cleavage of hybridized mRNA by the RNase P enzyme. Genes encoding for this U5-EGS (560) molecule, as well as a U5 EGS (560D) antisense control, were cloned into retroviral plasmids and transferred into a CD4+ T cell line. Transfected cells were exposed to increasing concentrations of HIV-1 clinical isolates from clades A, B, C, and F. Heterogeneous cultures of CD4+ T cells expressing the U5 EGS (560) molecule were observed to maintain CD4 levels, were devoid of cytopathology, and did not produce HIV p24 gag antigen through 30 days after exposure to all HIV-1 clades at a multiplicity of infection of 0.01. Identical cells expressing the U5 EGS (560D) antisense control molecule underwent a loss of CD4 expression, produced elevated levels of HIV-1, and formed large syncytia similar to untreated cells. When the viral inoculum was increased at the time of exposure (multiplicity of infection = 0.05), the inhibitory effect of the U5 EGS (560) molecule was overwhelmed, but viral-mediated cytopathology and particle production were delayed compared with control cell populations. Viral replication and cytopathology associated with infection of multiple HIV-1 clades can be effectively inhibited in CD4+ cells expressing the RNase P-associated U5 EGS (560) molecule.
RNA-based gene interference strategies have been proposed as useful research tools and potential therapeutic interventions for human malignancies and infectious diseases (1). These technologies include antisense oligonucleotides (2–4), ribozymes (5–10), RNase P-associated EGS molecules (EGS) (11–15), and, more recently, double-stranded interference RNA (RNAi) (16–19). Each of these strategies maintain inherent advantages and limitations with regard to target specificity, potency, and optimal expression and delivery in vivo (13, 20, 21). To date, there are several clinical trials evaluating antisense and ribozyme technologies targeting oncogenic or viral mRNA transcripts with potential therapeutic effects (22). However, since its discovery by Sidney Altman in the late 1970s and extensive characterization in bacterial systems, there are only a limited number of studies examining the use of sequence-specific, RNase P-associated EGS molecules to target disease-associated mRNA transcripts (23).
RNase P, a ribonucleoprotein that consists of enzyme and RNA subunits, catalyzes the hydrolysis reaction that removes the 5′ leader sequence from tRNA precursors to form mature tRNA molecules (24, 25). This essential enzymatic activity has been found in all cell types, including prokaryotes and eukaryotes (26). A general strategy for gene targeting has been developed based on the fact that the 3′ proximal sequence of a tRNA precursor molecule, called the external guide sequence (EGS), can be modified to hybridize with complementary target mRNA (11, 13, 14). The EGS is designed to leave a 5′-ACCAC-3′ unpaired stretch required for cleavage by the RNase P protein component following hybridization. In principle, any mRNA of known sequence can be targeted for precise cleavage by RNase P through use of custom designed EGS RNA without the restriction of a specific nucleotide sequence in the target mRNA (14, 27). This property permits construction of the EGS molecule to avoid areas of complex secondary structure that would severely limit the binding of ribozymes. Furthermore, unlike ribozymes, single point mutations within the target mRNA would not prevent recognition by the EGS and cleavage by RNase P (20). Subsequent studies of substrate recognition and RNase P cleavage requirements have provided insight into structural modifications of the RNA component of RNase P that may be implemented to enhance target mRNA cleavage (12). As such, RNase P molecules combine the attributes of antisense technologies and ribozymes, and transcend several of their limitations.
In a simple preliminary set of experiments, we designed and identified a truncated RNase P-associated EGS molecule [U5 EGS (560)] that specifically recognizes and hybridizes to the U5 region of the 5′ leader sequence of HIV-1 (ref. 28). This region is highly conserved across HIV-1 clades and is present on all forms of HIV RNA including incoming RNA strands and mRNA transcripts expressed following infection (7–10). We observed the long-term RNase P-mediated inhibition of HIV-1 infection, replication, and cytopathology by using a common laboratory strain of HIV-1 (Clade B) (28). The promising results from these preliminary experiments provided the basis for a comprehensive investigation of the inhibitory properties of this RNase P-associated EGS molecule by using clinical HIV-1 isolates from multiple clades to establish the potential application of this strategy for therapeutic applications. In the following report, we examine the capacity of this potent U5 EGS (560) molecule to protect CD4+ cells from infection and replication by various clades of HIV-1 clinical isolates. In addition, we evaluate the inhibitory properties of the U5 EGS (560) molecule as CD4+ cells are exposed to higher concentrations of HIV-1 clinical isolates. Finally, we begin to investigate at what point of the HIV-1 replication cycle this targeted RNase P-associated U5 EGS molecule mediates inhibition of viral infection.
Materials and Methods
Clinical HIV-1 Isolates Preparation of Viral Stocks.
HIV-1 clinical isolates corresponding to clades A, B, C, and F were contributed by the Joint United Nations Programme on HIV/AIDS Network for HIV Isolation and Characterization, and the Division of Acquired Immuno-Deficiency Syndrome, National Institutes of Health; and obtained from the National Institutes of Health AIDS Research and Reference Reagent Program.
Cell Culture.
M4C8 is a CD4+ T cell line permissive to both HIV-1 and HIV-2 (29). M4C8 were the kind gift of M. Hayami, Institute for Virus Research, Kyoto University, Kyoto.
RNase P-Associated EGS Molecule Design and Vector Construction.
The Moloney murine leukemia virus (MoMLV) vectors pLNL-6-U5 EGS (560) and pLNL-6-U5 EGS (560D) were engineered by inserting a t-RNAval promoter-U5 EGS cassette into the unique NheI site located in the U3 region of the MoMLV 3′LTR as described in Hnatyszyn et al. (28).
Transfection and Selection of M4C8 Cells.
M4C8 cells were seeded at 105 cells per ml of media into uncoated T25 suspension flasks. Cells were transfected with the pLNL-6-U5 EGS (560) or (560D) control plasmid (2 μg), using Fugene (Roche Biochemicals) according to manufacturer's specifications. Transfected cells were selected with G418 (500 μg/ml of media). Seven days before infection with HIV-1 clinical isolates, selection media was removed from the transfected cells and replaced with normal culture media without G418.
Confirmation of Vector Expression by Using Real-Time Reverse Transcription (RT)-PCR.
Total RNA was isolated from M4C8, M4C8 + U5 EGS (560), and M4C8 + U5 EGS (560D) cells by using a High Pure RNA Isolation kit (Roche Diagnostics) and analyzed using real-time RT-PCR to confirm vector gene and U5 EGS expression (28).
Infection of Transfected and Control M4C8 Cells with Clinical Isolates of HIV-1.
Multiple replicates (4) of experimental [expressing the U5 EGS (560) or (560D)] and control M4C8 cells (105 cells in 500 μl of media) were placed in 1.5-ml microcentrifuge tubes. HIV-1 clinical isolates corresponding to clades A, B, C, and F (moi = 0.01 and 0.05) were added to each tube and incubated with the cells for 6 h. Following incubation, cells were washed five times with culture media by means of centrifugation to remove remaining virus. Cells were resuspended in 7 ml of fresh culture media and primary cell and supernatant samples were harvested for analysis (time = 0). Supernatants and cell samples were harvested from each culture at regular intervals over a period of 30 to 60 days for analysis for HIV infection, replication, and levels of CD4 expression.
CD4 Expression and Detection by Using Flow Cytometry.
Noninfected and infected cell samples were evaluated in triplicate for CD4 by using flow cytometry.
P24 Antigen Capture Assay for HIV-1 Replication.
HIV-1 production was monitored by determination of the p24 gag antigen concentration in the culture supernatants harvested at various time points, using the p24 Antigen Capture ELISA (Coulter p24 Antigen Assay, Beckman Coulter).
Single-Round Infection of Transfected and Control M4C8 Cells with HIV-1.
Transfected and control cells were exposed to a single concentration of HIV-1 and harvested at 12 h for PCR analysis to detect integrated proviral DNA. This time frame is sufficient for viral infection, reverse transcription, proviral integration, and limited viral replication to occur (30). Multiple replicates (4) of experimental (expressing the active or disabled U5 EGS) and control M4C8 cells (105 cells in 500 μl of media) were placed in 1.5-ml microcentrifuge tubes. An HIV-1 clinical isolate from Clade B (moi = 0.01) was added to each tube and incubated with the cells for 6 h. Following incubation, cells were washed five times with culture media by means of centrifugation to remove remaining virus.
Genomic DNA was isolated from cell samples harvested at 12 h postinfection, using a commercial kit (Perfect Blood gDNA Mini, Eppendorf). DNA was quantitated using spectrophotometry and analyzed for the presence of HIV-1 proviral DNA as an indication of infection using real-time PCR and the LightCycler. PCR mixtures were prepared according to the DNA Amplification SYBR Green I Kit (Roche Biochemicals) using the SK38 (5′-ATAATCCACCTATCCCAGTAGGAGAAAT-3′) and SK39 (5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′) oligonucleotides specific for the viral sequence encoding a region of HIV-1 gag protein. The real-time PCR conditions were 30 cycles at 95°C for 1 s, 60°C for 5 s, and 72°C for 20 s.
Results
Levels of RNase P-Associated U5 EGS Molecule Expression in a CD4+ T Cell Line Transfected with the Retroviral Plasmids.
The CD4+ T cell line, M4C8, was transfected with each of the retroviral plasmids expressing either the U5 EGS (560) or U5 EGS (560D) antisense control. Transfected cells were selected for NeoR by culturing the cells in culture media supplemented with G418. Single-cell clones were not generated from the NeoR population and the “bulk” culture for each RNase P-associated U5 EGS molecule was used to evaluate cross clade infectivity and cytopathology. We believe these heterogeneous cell populations contain cells that express both high and low levels of each U5 EGS molecule and would better represent variable gene expression that would occur in vivo.
Initially, the NeoR cell populations were evaluated for changes in cell proliferation compared with nontransfected M4C8 cells by using nonradioactive cell proliferation assays as a measure of potential toxicity induced by expression of the RNase P-associated EGS molecules. No significant alterations in the rates of cell division between nontransfected M4C8 cells and cells transfected with the retroviral plasmids expressing either the U5 EGS (560) or antisense control were observed (data not shown). Total RNA from each cell population was converted to cDNA and examined using quantitative real-time PCR to determine the average levels of expression of either the U5 EGS (560) or (560D) (Table 1). Using GAPDH as a control for levels of cDNA added to each PCR reaction, crossing point ratios for each RNase P-associated U5 EGS molecule as a measure of expression levels were generated. These ratios indicated that expression of the disabled U5 EGS (560D) was not significantly different from the U5 EGS (560) in each transfected M4C8 cell population. The levels of U5 EGS (560) and (560D) expression were determined to be similar in each selected M4C8 population used for this investigation and generated no overt adverse effects to the cells as measured by changes in cell viability and proliferation.
Table 1.
Detection of U5 EGS (560) and (560D) expression by using cDNA and real-time PCR
| Cellular RNA/cDNA | U5 EGS (Crossing Point) | GAPDH (Crossing Point) | U5 EGS/GAPDH Ratio |
|---|---|---|---|
| M4C8 | 0.00 | 19.52 | 0.000 |
| M4C8 + U5 EGS (560) | 18.30 | 15.94 | 1.148 |
| M4C8 + U5 EGS (560D) | 20.62 | 15.95 | 1.293 |
Cross-Clade Protection at a moi of 0.01.
The main objective of this investigation was to determine whether U5 EGS (560) molecules provided cross-clade protection from HIV-1 infection and replication, using a standard viral load. From our previous experiments with these anti-HIV-1 RNase P-associated U5 EGS molecules, we observed that M4C8 cells expressing the U5 EGS (560) were protected from infection and replication by a laboratory strain of HIV-1 (HIV-1 MN; moi = 0.01). In this investigation, we obtained HIV-1 clinical isolates corresponding to clades A, B, C, and F to evaluate the potency of this inhibition observed with the U5 EGS (560) but not the disabled form of the molecule. M4C8 cell populations expressing the U5 EGS (560) or the U5 EGS (560D) antisense control, as well as nontransfected M4C8 cells, were exposed to the HIV-1 isolates from each viral clade (moi = 0.01). Cell and supernatant samples were harvested from each of the cultures at regular time intervals for analysis to determine HIV infection and replication. Each culture was also examined using light microscopy on a daily basis to detect the formation of cell syncytia as an indication of HIV cytopathology.
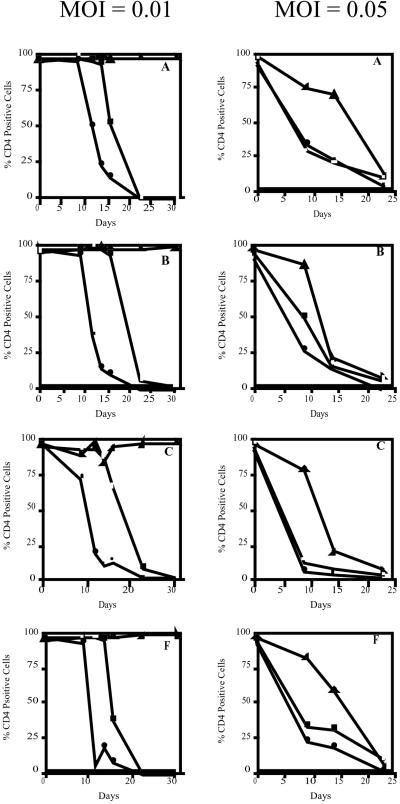
Nontransfected M4C8 were permissive to infection by all four clades of HIV-1. Levels of CD4 expression remained steady in this control cell population for 16–18 days postinitial exposure to each viral clade at a moi of 0.01 (Fig. 1). At this time, analysis by flow cytometry indicated a significant decline in CD4 expression in the cell population with a complete loss of CD4 cells by 23 days postexposure. Syncytia formation was first noted in these control cultures at 12–14 days postinfection and was significant by 18–20 days postinfection. At 24 days postexposure to each HIV-1 clade, less cytopathology was observed in these cultures and CD4−, HIV-1-infected M4C8 clones began to dominate the culture environment.
Figure 1.
CD4 expression by nontransfected M4C8 cells and cells expressing the RNase P associated U5 EGS molecules following exposure to HIV-1 clades A, B, C, and F (moi = 0.01 and 0.05). At a moi of 0.01 and 0.05, there was a reduction in CD4 expression by nontransfected M4C8 and M4C8 expressing the U5 EGS (560D) antisense control following exposure to each clade of HIV-1. The percentage of CD4 expressing cells remained constant in cultures of M4C8 cells expressing the U5 EGS (560) molecule following exposure to each HIV-1 clade at a moi of 0.01. At the elevated moi, viral mediated cytopathology was delayed in CD4+ cells expressing RNase P-associated U5 EGS (560) molecules. □, M4C8; ▴, M4C8 + U5 EGS (560); ●, M4C8 + U5 EGS (560D).
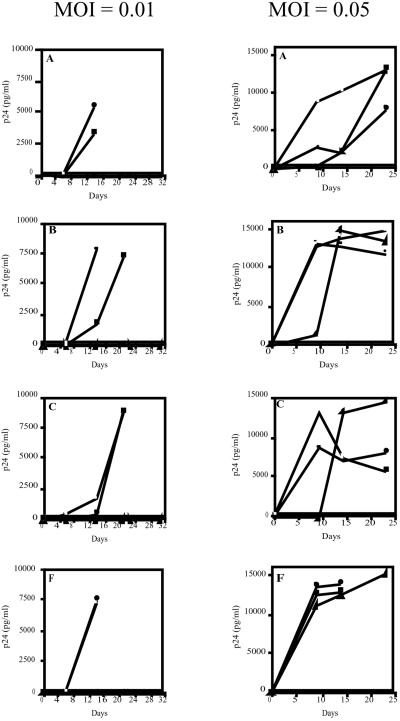
The loss of CD4 and syncytia formation correlated with increased levels of HIV-1 p24 antigen as an indication of HIV replication and production (Fig. 2). At 14 days postexposure to each clade of HIV-1, all nontransfected M4C8 culture supernatant had significant levels of p24 antigen (1,800–7,500 pg/ml). Supernatants from M4C8 cells exposed to HIV-1 clinical isolates from clades B and C had slightly lower levels of p24 antigen. These lower levels of viral production may be attributed to variations of infectivity and pathogenesis within each HIV-1 clade or to experimental deviations in determining each moi. By 20–23 days postinfection, p24 antigen levels were very high in all cultures of nontransfected M4C8 control cells (<8,000 pg/ml). These results indicate that nontransfected M4C8 cells are permissive to infection by each of the HIV-1 clades examined, undergo viral-mediated cytopathology, and produce high levels of virus following exposure to a standard moi of each HIV-1 isolate.
Figure 2.
HIV-1 p24 antigen levels in supernatants from nontransfected M4C8 cells and cells expressing the RNase P-associated U5 EGS Molecules and exposed to HIV-1 clades A, B, C, and F (moi = 0.01 and 0.05). □, M4C8; ▴, M4C8 + U5 EGS (560); ●, M4C8 + U5 EGS (560D).
M4C8 cells expressing the U5 EGS (560D) antisense control molecule were also permissive to all of the HIV-1 clades tested. As observed in our previous study when using HIV-1 MN, cells expressing the disabled form of the RNase P-associated U5 EGS molecule appear to be more susceptible to HIV-1 infection and cytopathology (28). Significant loss of CD4 expression was noted by 9–12 days postexposure to each viral clade and syncytia formation was recorded at 5–7 days (Fig. 1). Similar to nontransfected M4C8 cells exposed to each HIV-1 clade, detectable syncytia in cultures of M4C8 cells expressing the U5 EGS (560D) dissipated around 24 days postexposure and the formation of CD4−, HIV-1-infected clones were prominent in the viable cell population.
HIV-1 p24 antigen levels were significantly elevated in culture supernatants at 14 days post exposure for HIV-1 clades A, B, and F (5,400–7,900 pg/ml) with lower but detectable levels in cells exposed to clade C (600 pg/ml; Fig. 2). By 21 days postexposure, supernatant from M4C8 cells expressing the U5 EGS (560D) antisense control and exposed to the HIV-1 isolate from clade C had levels of p24 antigen above 8,000 pg/ml. These observations indicate that expression of the disabled U5 EGS (560D) molecule does not have a measurable effect on the permissivity of M4C8 cells to infection with each clade of HIV-1 and does not inhibit viral infection, replication, or cytopathology.
In contrast, there was no loss of CD4 expression or detectable syncytia formation in M4C8 cells expressing the U5 EGS (560) following exposure to each of the HIV-1 isolates (Fig. 1). Furthermore, there were no detectable levels of HIV-1 p24 in the culture supernatant through 30 days postexposure to each clade of HIV-1 (Fig. 2). These cultures were maintained past 60 days postexposure and there was no loss of CD4 or measurable p24 antigen in culture supernatants harvested from M4C8 cells expressing the U5 EGS (560) molecule. These results suggest that M4C8 cells expressing the RNase P-associated U5 EGS (560) molecule are protected from infection, replication, and subsequent cytopathology following exposure to multiple clades of HIV-1 at a significant viral load (moi = 0.01).
Increasing moi Reduces EGS Inhibition.
We evaluated the protective capacity of the RNase P-associated U5 EGS (560) molecule when M4C8 cells expressing this molecule were exposed to higher viral loads of each HIV-1 clinical isolate. Nontransfected M4C8 cells and M4C8 cells expressing either the U5 EGS (560) of U5 EGS (560D) antisense control molecule were exposed to each HIV-1 clinical isolate at concentrations 5 times the original viral load (moi = 0.05). Cell and supernatant samples were harvested at regular intervals to examine viral infection and production. Light microscopy was used to monitor HIV-1 exposed cultures for the formation of syncytia as an indication of viral-mediated cytopathology.
Nontransfected M4C8 cells, as well as M4C8 cells expressing the disabled U5 EGS (560D) molecule, yielded similar results following exposure to the higher viral loads. For each HIV-1 isolate, there was a significant loss of CD4 expression by 9 days postexposure for all cultures at a moi of 0.05 (Fig. 1). The loss of CD4 expression by these cells at this time point ranged from 51–92%. By 14 days post exposure to each HIV-1 clade, cultures of both nontransfected M4C8 and cells expressing the U5 EGS (560D) antisense control molecule had less than 30% CD4+ cells. In general, syncytia formation in these cultures was observed at 4 days postexposure and was significant in each population at 9 days postinfection with each clade. Furthermore, there were no viable cells observed in cultures of nontransfected M4C8 or M4C8 expressing the U5 EGS (560D) molecule and exposed to HIV-1 clade F by 16 to 18 days postexposure. Indeed, each of these control cultures exposed to the elevated moi from each viral clade had no viable cells by 24 to 30 days postinfection.
Analysis of culture supernatants for HIV-1 p24 antigen levels as a measure of virus particle production revealed significant levels (2,500–13,900 pg/ml) of the viral antigen at 9 days postexposure to each HIV-1 clade (Fig. 2). At subsequent time points, the level of p24 antigen in each culture supernatant was variable and depended on the number of viable cells remaining in culture. For example, at 14 to 23 days postexposure to HIV-1 clades A, B, and F, the levels of p24 antigen in the supernatants harvested from nontransfected M4C8 and cells expressing the U5 EGS (560D) antisense control molecule were elevated (<10,000 pg/ml) compared with earlier time points. However, at the same time points, HIV-1 p24 antigen levels for similar cells exposed to clade C either declined or stabilized because of a lower number of viable cells in culture producing HIV-1 particles. Indeed, in cultures of nontransfected M4C8 cells and cells expressing the U5 EGS (560D) molecule and infected with HIV-1 clade F, there were no viable cells observed past 16 days postexposure and p24 was not measured past that time (Fig. 2). As expected, exposure of nontransfected M4C8 cells or cells expressing the U5 EGS (560D) antisense control molecule to higher viral loads resulted in accelerated viral-mediated cytopathology and increased viral production compared with the standard moi of 0.01.
M4C8 cells expressing the RNase P-associated U5 EGS (560) molecule were permissive to infection by each HIV-1 isolate at a moi of 0.05. However, the depletion of CD4 expression and syncytia formation was significantly delayed in this cell population following exposure to the higher viral load (Fig. 1). M4C8 cells expressing the U5 EGS (560) molecule and exposed to the HIV-1 clades A, B, C, and F had a small, but detectable loss of CD4 expression at 9 days postexposure (12–24%). For cells exposed to HIV-1 clade A, there was no further significant decline of CD4 levels until 14 to 23 days postinfection. Cells exposed to clades B, C, and F had noticeable losses of CD4+ cells by 14 days postinfection. For all cultures at this elevated moi, there was less than 15% CD4+ cells by 23 days postexposure. Syncytia formation was initially detectable in M4C8 cells expressing the U5 EGS (5,600 molecule at 6 to 8 days postexposure to HIV-1 clades A, B, C and F). However, abundant syncytia were not observed in any of the cultures until 12 to 14 days postinitial exposure with significantly less cytopathology in cultures infected with HIV-1 clade A. In addition, viable cells were observed in all cultures of M4C8 cells expressing the RNase P-associated U5 EGS (560) molecule through 24 days postexposure, but all cultures eventually succumbed to infection by 30–36 days postinfection.
At a moi of 0.05, the appearance of HIV-1 p24 antigen in each culture supernatant varied with each viral clade following exposure of M4C8 cells expressing the U5 EGS (560) molecule (Fig. 2). At 9 days postexposure to HIV-1 clades A, B, and C, either no or very low levels of p24 antigen was detectable in the culture supernatants of M4C8 cells expressing the U5 EGS (560) molecule. However, similar cells exposed to the HIV-1 clinical isolate from clade F had significant p24 antigen levels (<10,000 pg/ml) at that same time point. By 14 days post exposure to each HIV-1 clade, the p24 antigen levels were significantly increased and continued to elevate through 23 days postinfection. These results indicate that CD4+ cells expressing the RNase P-associated U5 EGS (560) molecule can be infected by various clades of HIV-1 at a higher viral load. However, cells expressing the U5 EGS (560) molecule are afforded limited protection from HIV-1 infection and replication as indicated by a delay in loss of CD4 expression, syncytia formation, and viral production compared with control cultures.
Single-Round Infection Experiment.
Based on our preliminary experiments and the results from this investigation, M4C8 cells expressing the RNase P-associated U5 EGS (560) molecule do not undergo typical cytopathology following exposure to moderate titers of HIV-1 (moi = 0.01). However, we have had little insight into the point of the HIV replication cycle where this inhibition by the RNase P-associated EGS molecule occurs. We designed a time-restricted experiment that examined the presence of proviral DNA in M4C8 cells following exposure to HIV-1 MN at a moi of 0.01. The duration of the experiment was limited to 12 h postexposure of cells to HIV-1. This restricted period is roughly equivalent to the time required for a virion to complete a single round of the viral replication cycle from infection to integration and subsequent production. We used real-time PCR with genomic DNA isolated from M4C8 expressing the U5 EGS (560) and U5 EGS (560D) antisense control to detect integrated proviral DNA. The presence or absence of proviral DNA in each sample would provide insight into the point of inhibition by the U5 EGS (560) molecule.
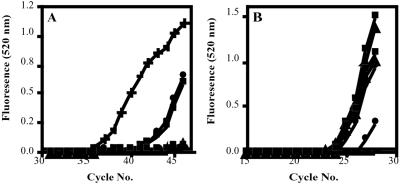
All genomic DNA samples were analyzed for the presence of the B7 gene to control for levels of genomic DNA isolated from each M4C8 cell population (Fig. 3B). For each sample, crossing points corresponding to the amplification of the B7 internal control and detectable HIV-1 proviral DNA would be used to determine ratios for comparison of the presence of integrated HIV genome (Table 2). HIV-1 proviral DNA was readily detectable in genomic DNA samples isolated from nontransfected M4C8 cells and cells expressing the U5 EGS (560D) molecule at 12 h postexposure (Fig. 3A; Table 2). These results would indicate that HIV-1 infection, reverse transcription, and integration were not inhibited as proviral DNA was integrated into the host cell genome. These observations would correlate with our previous results where nontransfected M4C8 cells and cells expressing the U5 EGS (560D) antisense control molecule were readily infected with HIV-1 and underwent viral-mediated cytopathology. In contrast, HIV-1 proviral DNA was not detectable by real-time PCR in genomic DNA harvested from M4C8 cells expressing the U5 EGS (560) molecule at 12 h postexposure. Once again, the lack of evidence for proviral DNA in these CD4+ cells expressing the RNase P-associated U5 EGS (560) molecule corresponds with the results from our experiments where M4C8 cells expressing the U5 EGS (560) molecule were protected from viral-mediated cytopathology and replication. Although it is possible that real-time PCR may not have the sensitivity to detect a single copy of the integrated HIV-1 genome, these results would indicate that at least some of the inhibitory effects attributed to the U5 EGS (560) molecule occur before proviral DNA synthesis. This inhibition would most likely occur before reverse transcription of the incoming viral RNA.
Figure 3.
Real-time PCR detection of HIV-1 proviral DNA in genomic DNA isolated from nontransfected M4C8 cells and cells expressing the RNase P-associated U5 EGS molecules following exposure to HIV-1 (moi = 0.01) at 12 h postinfection. (A) Real-time PCR amplification curves for genomic DNA samples to detect the presence of proviral HIV-1 DNA. (B) Real-time PCR amplification curves for genomic DNA samples to detect the internal control B7 gene. ×, Negative control; +, HIV-1 DNA; ▵, M4C8 + U5 EGS (560) 0 h; ▴, M4C8 + U5 EGS (560) 12 h; □, M4C8 0 h; ■, M4C8 12 h; ○, M4C8 + U5 EGS (560D) 0 h; ●, M4C8 + U5 EGS (560D) 12 h.
Table 2.
Summary of real-time PCR results for the detection of HIV-1 proviral DNA in genomic DNA isolated from nontransfected M4C8 cells and cells expressing the RNase P-associated U5 EGS molecules and exposed to HIV-1 (moi = 0.01) at 12 h postinfection
| Cell line | HIV-1 crossing point
|
B7 crossing point
|
HIV-1/B7 ratio
|
|||
|---|---|---|---|---|---|---|
| 0 hr | 12 hr | 0 hr | 12 hr | 0 hr | 12 hr | |
| M4C8 | 0.00 | 42.62 | 22.22 | 23.86 | 0.000 | 1.786 |
| M4C8 + U5 EGS (560) | 0.00 | 0.00 | 22.54 | 23.59 | 0.000 | 0.000 |
| M4C8 + U5 EGS (560D) | 0.00 | 42.06 | 24.02 | 26.20 | 0.000 | 1.605 |
Discussion
The RNase P complex may offer an excellent alternative to conventional RNA-based gene interference therapies for the treatment of infectious diseases and human malignancies. The RNase P-associated EGS molecule can be designed to recognize and hybridize to any target mRNA regardless of target sequence motifs. The enzyme component of RNase P required for cleavage of the RNA hybrid is ubiquitously expressed in all cells that express tRNA and translate proteins. The result is a specific, effective, and versatile gene interference system that is independent of the target mRNA sequence and can be applied in every cell type.
In this investigation, we evaluated an RNase P-associated EGS molecule targeting the U5 region of the 5′ leader sequence of HIV-1. Preliminary experiments suggested that a CD4+ T cell line expressing the U5 EGS (560) molecule was protected from infection by a common laboratory strain of HIV-1 at a moi of 0.01 (28). Based on localization studies suggesting a nuclear locale for the RNase P enzyme, we hypothesized that inhibition of viral infection and subsequent cytopathology by the U5 EGS (560) molecule could occur at the level of incoming viral RNA, as well as RNA transcribed from integrated HIV-1 proviral DNA (5, 31–33).
The results of this new set of experiments has provided insight into the effectiveness of the RNase P-associated U5 EGS (560) molecule against increasing concentrations of clinical isolates of HIV-1 from multiple clades. Following exposure to a standard moi of 0.01 from HIV-1 isolates of clades A, B, C, and F, M4C8 cells expressing the U5 EGS (560) molecule were protected from viral-mediated cytopathology and viral replication. Nontransfected M4C8 and cells expressing the U5 EGS (560D) antisense control molecule were infected with each viral clade, underwent loss of CD4 and syncytia formation, and generated viral progeny. These results indicate that the viral inhibition attributed to the U5 EGS (560) molecule is not due to an antisense effect, as the disabled molecule shares the same recognition sequence as the inhibitory form. Thus, cells expressing the U5 EGS (560) molecule were protected from cross-clade HIV-1 infection and cytopathology by RNase P-mediated inhibition. These promising results were not unexpected because the RNase P-associated U5 EGS (560) molecule was designed to recognize a region of the HIV-1 genome that is highly conserved across viral clades and is not prone to mutation. However, as observed with other RNA-based gene interference strategies, secondary structure of viral RNA may vary between HIV-1 clades and accessibility of the target sequence may affect the efficacy of the U5 EGS molecule (21). We did not observe any such restrictions of the U5 EGS (560) molecule to target accessibility within each of the HIV-1 clinical isolates examined in this study.
We also evaluated the protective capacity of the RNase P-associated U5 EGS (560) molecule in cells when they are exposed to higher viral loads. Based on theoretical calculations using the number of circulating CD4+ cells in healthy individuals and the average viral load of an HIV patient, a moi of 0.01 is similar to the potential level of exposure of a human being to infectious HIV-1 in vivo (34). When exposed to 5 times the standard moi, cells expressing the RNase P-associated U5 EGS (560) molecule experienced a cross-clade delay in the onset of viral cytopathology, loss of CD4 expression, and, to some extent, viral production compared with control cell cultures. These results suggest that the inhibitory effects of the U5 EGS (560) molecule can be reduced by exposure to high titers of HIV-1 clinical isolates. This observation is predictable and expected because the therapeutic effect of most antiviral drugs and other RNA-based interventions for HIV-1 can be overwhelmed by initial exposures to high titers of virus (35–37). Furthermore, our transfected cell cultures are heterogeneous for levels of RNase P-associated U5 EGS molecule expression. Therefore, a portion of the cell population with very low levels of protective U5 EGS (560) expression serve as sources for viral replication eventually increasing the titer of infectious virus to overwhelm cells that express high levels of the inhibitory molecule. Indeed, the delay in onset of viral mediated cytopathology in cells expressing the RNase P-associated U5 EGS (560) molecule would suggest that methods to improve expression of the U5 EGS (560) or a combination with other forms of HIV-1 interventions could protect cells from exposure to these high viral loads. We are currently evaluating vector systems that can deliver multiple expression cassettes encoding for the U5 EGS (38).
Finally, we used real-time PCR and a time-restricted single-round infection experiment to gain information regarding the site of U5 EGS (560) inhibition within the HIV-1 replication cycle. Nontransfected M4C8 cells and cells expressing either the U5 EGS (5600) or (560D) molecules were exposed to HIV-1 MN at a moi of 0.01. At 12 h postexposure, the approximate time required to achieve viral entry, reverse transcription, viral integration, and early transcription, cells were harvested and genomic DNA was isolated for analysis. Real-time PCR was used to detect the presence of integrated HIV-1 proviral DNA in each sample. Although HIV-1 proviral DNA was readily detectable in DNA isolated from nontransfected M4C8, as well as cells expressing the U5 EGS (560D) antisense control molecule, proviral DNA was not detected in DNA samples from M4C8 cells expressing the RNase P-associated U5 EGS (560) molecule. These results would indicate, at least in part, that the U5 EGS (560) molecule inhibits the HIV-1 replication cycle at the level of incoming viral RNA, before or during reverse transcription. This observation has significance regarding the use of the RNase P associated U5 EGS molecules for HIV intervention because most of the current antiviral approaches have difficulty preventing HIV proviral integration, leading to the presence of latent viral reservoirs. However, the mechanism of this viral inhibition by the U5 EGS (560) molecule before viral integration remains unclear. Our PCR results must be tempered with the possibility that real-time PCR may not have the sensitivity to detect a limited number of integrated proviral HIV-1 genomes and that U5 EGS (560) inhibition of viral replication still occurs postintegration at the level of viral genome transcription. Another possibility would have the U5 EGS transcript transported to the cytoplasm of transfected cells following expression by the tRNA promoter and recruiting the RNase P enzyme to inhibit the generation of proviral DNA before reverse transcription of incoming viral RNA (5, 33). Alternatively, RNase P-associated EGS molecules may bind to the U5 region of incoming HIV RNA and interfere with the initiation or completion of reverse transcription. This scenario would not require the recruitment of the RNase P enzyme for cleavage of the RNA hybrid. However, we would expect similar inhibition to occur in cells expressing the disabled RNase P associated EGS molecule because it shares an identical hybridization region with the U5 EGS (560) molecule and this was not observed. It is quite possible that the U5 EGS (560) molecule can bind and mediate RNase P cleavage of both incoming viral RNA and viral transcripts generated following proviral integration. Further investigation regarding the intracellular distribution of the RNase P enzyme component and RNase P associated EGS molecules following expression by pol III promoters will further clarify the site of site of HIV inhibition. However, we believe that this single-round infection experiment confirms inhibition by the RNase P-associated U5 EGS (560) molecule at the level of incoming viral RNA. These results would account for the undetectable levels of proviral DNA in exposed cell populations and is supported by the lack of viral antigen detection in long-term culture.
These early investigations examining the inhibitory effects of the RNase P-associated U5 EGS (560) molecule have yielded promising results with regard to the cross-clade protection and potency of RNase P-mediated inhibition of HIV-1 infection. Further experimentation will involve improving expression of the U5 EGS (560) molecule, combining this molecule with other potent HIV-1 inhibitors and evaluation of this molecule in primary CD4+ cell populations and hematopoietic stem cells. Future directions could also include the production of synthetic RNase P-associated EGS molecules that could be administered much like pharmaceuticals or in combination with targeted drug delivery systems as an effective intervention for HIV infection (39).
Abbreviations
- EGS
external guide sequence
- moi
multiplicity of infection
- M4C8
MOLT 4 Clone 8 CD4+ T cell line
- NeoR
neomycin resistance
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wong-Staal F, Buchschacher G L., Jr Gene Ther. 2000;7:1351–1352. doi: 10.1038/sj.gt.3301267. [DOI] [PubMed] [Google Scholar]
- 2.Chadwick D R, Lever A M. Gene Ther. 2000;7:1362–1368. doi: 10.1038/sj.gt.3301254. [DOI] [PubMed] [Google Scholar]
- 3.Giles R V, Tidd D M. Nucleic Acids Res. 1992;20:763–770. doi: 10.1093/nar/20.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giles R V, Ruddell C J, Spiller D G, Green J A, Tidd D M. Nucleic Acids Res. 1995;23:954–961. doi: 10.1093/nar/23.6.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato Y, Kuwabara T, Warashina M, Toda H, Taira K. J Biol Chem. 2001;276:15378–15385. doi: 10.1074/jbc.M010570200. [DOI] [PubMed] [Google Scholar]
- 6.Klebba C, Ottmann O G, Scherr M, Pape M, Engels J W, Grez M, Hoelzer D, Klein S A. Gene Ther. 2000;7:408–416. doi: 10.1038/sj.gt.3301094. [DOI] [PubMed] [Google Scholar]
- 7.Leavitt M C, Yu M, Yamada O, Kraus G, Looney D, Poeschla E, Wong-Staal F. Hum Gene Ther. 1994;5:1115–1120. doi: 10.1089/hum.1994.5.9-1115. [DOI] [PubMed] [Google Scholar]
- 8.Ojwang J O, Hampel A, Looney D J, Wong-Staal F, Rappaport J. Proc Natl Acad Sci USA. 1992;89:10802–10806. doi: 10.1073/pnas.89.22.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada O, Yu M, Yee J K, Kraus G, Looney D, Wong-Staal F. Gene Ther. 1994;1:38–45. [PubMed] [Google Scholar]
- 10.Yamada O, Kraus G, Leavitt M C, Yu M, Wong-Staal F. Virology. 1994;205:121–126. doi: 10.1006/viro.1994.1626. [DOI] [PubMed] [Google Scholar]
- 11.Altman S. Proc Natl Acad Sci USA. 1993;90:10898–10900. doi: 10.1073/pnas.90.23.10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma M Y, Jacob-Samuel B, Dignam J C, Pace U, Goldberg A R, George S T. Antisense Nucleic Acid Drug Dev. 1998;8:415–426. doi: 10.1089/oli.1.1998.8.415. [DOI] [PubMed] [Google Scholar]
- 13.Ma M, Benimetskaya L, Lebedeva I, Dignam J, Takle G, Stein C A. Nat Biotechnol. 2000;18:58–61. doi: 10.1038/71924. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Hwang E S, Altman S. Proc Natl Acad Sci USA. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawa D, Wang J, Yuan Y, Liu F. RNA. 1998;4:1397–1406. doi: 10.1017/s1355838298980918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carthew R W. Curr Opin Cell Biol. 2001;13:244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- 18.Lin S L, Chuong C M, Ying S Y. Biochem Biophys Res Commun. 2001;281:639–644. doi: 10.1006/bbrc.2001.4412. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 20.Cobaleda C, Sanchez-Garcia I. Blood. 2000;95:731–737. [PubMed] [Google Scholar]
- 21.Cobaleda C, Sanchez-Garcia I. Trends Biotechnol. 2001;19:406–411. doi: 10.1016/S0167-7799(01)01741-3. [DOI] [PubMed] [Google Scholar]
- 22.Lamothe B, Joshi S. Front Biosci. 2000;5:D527–D555. doi: 10.2741/lamothe. [DOI] [PubMed] [Google Scholar]
- 23.Altman S. Nat Struct Biol. 2000;7:827–828. doi: 10.1038/79566. [DOI] [PubMed] [Google Scholar]
- 24.Altman S, Baer M F, Bartkiewicz M, Gold H, Guerrier-Takada C, Kirsebom L A, Lumelsky N, Peck K. Gene. 1989;82:63–64. doi: 10.1016/0378-1119(89)90030-9. [DOI] [PubMed] [Google Scholar]
- 25.Altman S, Kirsebom L, Talbot S. FASEB J. 1993;7:7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- 26.Altman S, Wesolowski D, Puranam R S. Genomics. 1993;18:418–422. doi: 10.1006/geno.1993.1488. [DOI] [PubMed] [Google Scholar]
- 27.Yuan Y, Altman S. EMBO J. 1995;14:159–168. doi: 10.1002/j.1460-2075.1995.tb06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hnatyszyn, H. J., Spruill, G., Young, A. K., Seivright, R. & Kraus, G. (2001) Gene Ther., in press. [DOI] [PubMed]
- 29.Kikukawa R, Koyanagi Y, Harada S, Kobayashi N, Hatanaka M, Yamamoto N. J Virol. 1986;57:1159–1162. doi: 10.1128/jvi.57.3.1159-1162.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada O, Kraus G, Luznik L, Yu M, Wong-Staal F. J Virol. 1996;70:1596–1601. doi: 10.1128/jvi.70.3.1596-1601.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartkiewicz M, Gold H, Altman S. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson M R, Cao L G, Taneja K, Singer R H, Wang Y L, Pederson T. J Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- 33.Kuwabara T, Warashina M, Sano M, Tang H, Wong-Staal F, Munekata E, Taira K. Biomacromolecules. 2001;2:1229–1242. doi: 10.1021/bm0101062. [DOI] [PubMed] [Google Scholar]
- 34.Hirschel B. Primary HIV Infection. London: Harcourt; 1999. [Google Scholar]
- 35.Jensen-Fangel S, Kirk O, Blaxhult A, Gerstoft J, Pedersen C, Black F T, Lundgren J D, Obel N. HIV Clin Trials. 2001;2:122–127. doi: 10.1310/KED4-8TW3-4ARG-HFC5. [DOI] [PubMed] [Google Scholar]
- 36.Fitzgibbon J E, Gaur S, Walsman S M, Janahi M, Whitley-Williams P, John J F., Jr AIDS Res Hum Retroviruses. 2001;17:1321–1328. doi: 10.1089/08892220152596579. [DOI] [PubMed] [Google Scholar]
- 37.Servais J, Nkoghe D, Schmit J C, Arendt V, Robert I, Staub T, Moutschen M, Schneider F, Hemmer R. J Acquired Immune Defic Syndr. 2001;28:221–225. doi: 10.1097/00042560-200111010-00003. [DOI] [PubMed] [Google Scholar]
- 38.Gervaix A, Li X, Kraus G, Wong-Staal F. J Virol. 1997;71:3048–3053. doi: 10.1128/jvi.71.4.3048-3053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hnatyszyn H J, Kossovsky N, Gelman A, Sponsler E. PDA J Pharm Sci Technol. 1994;48:247–254. [PubMed] [Google Scholar]