Abstract
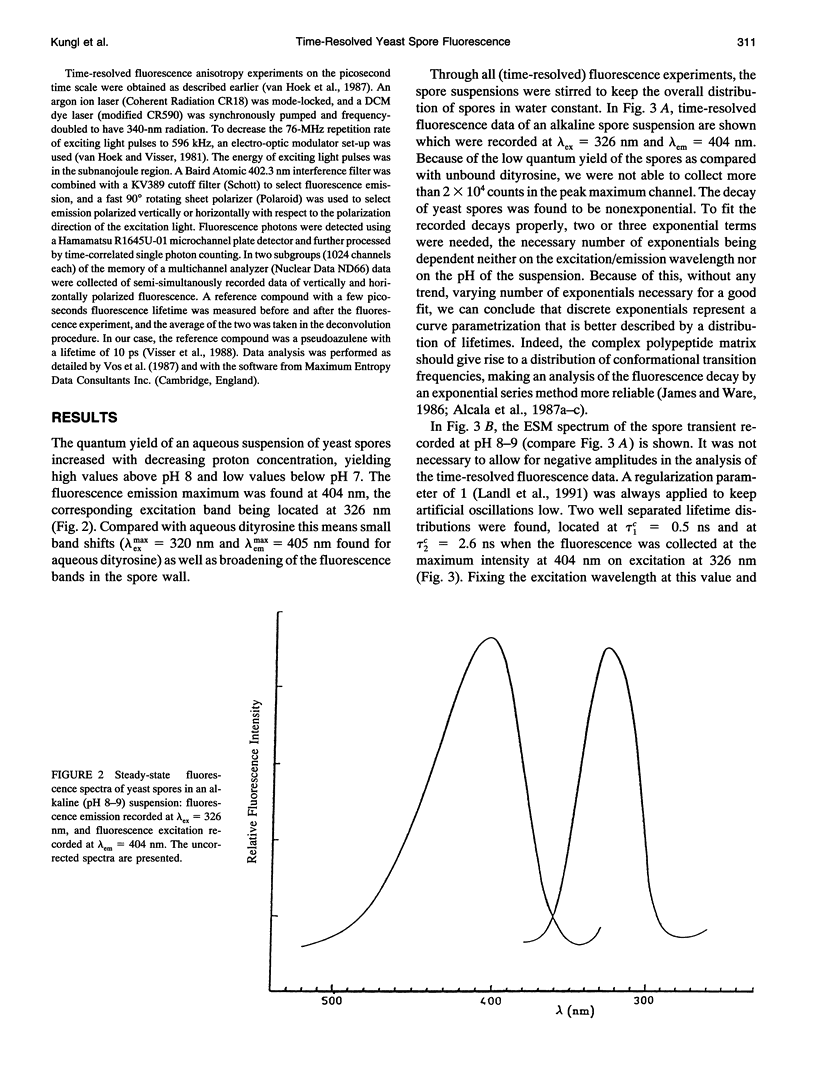
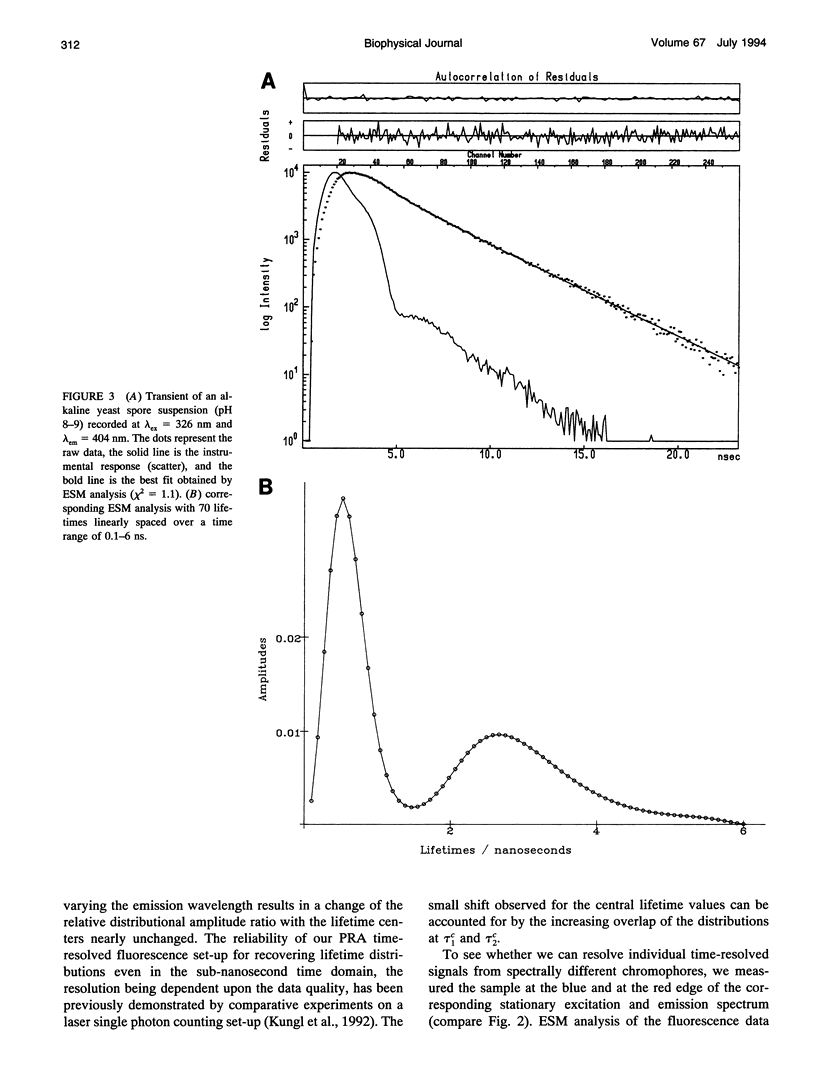
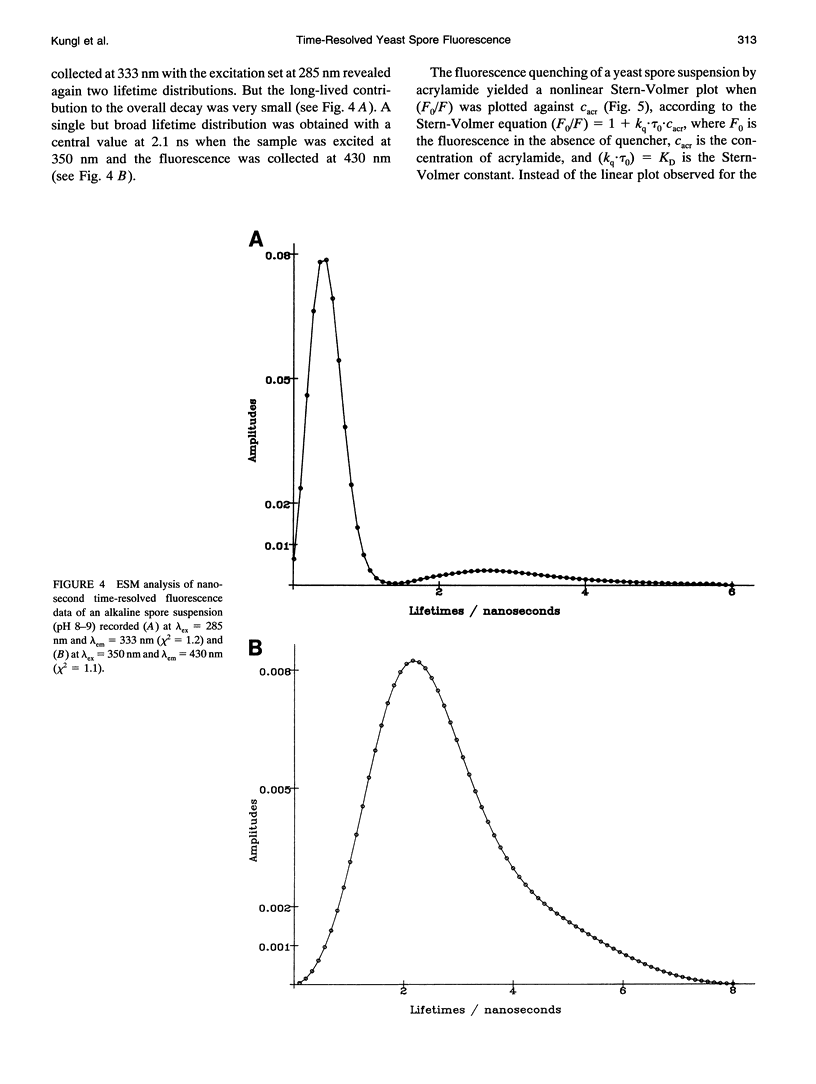
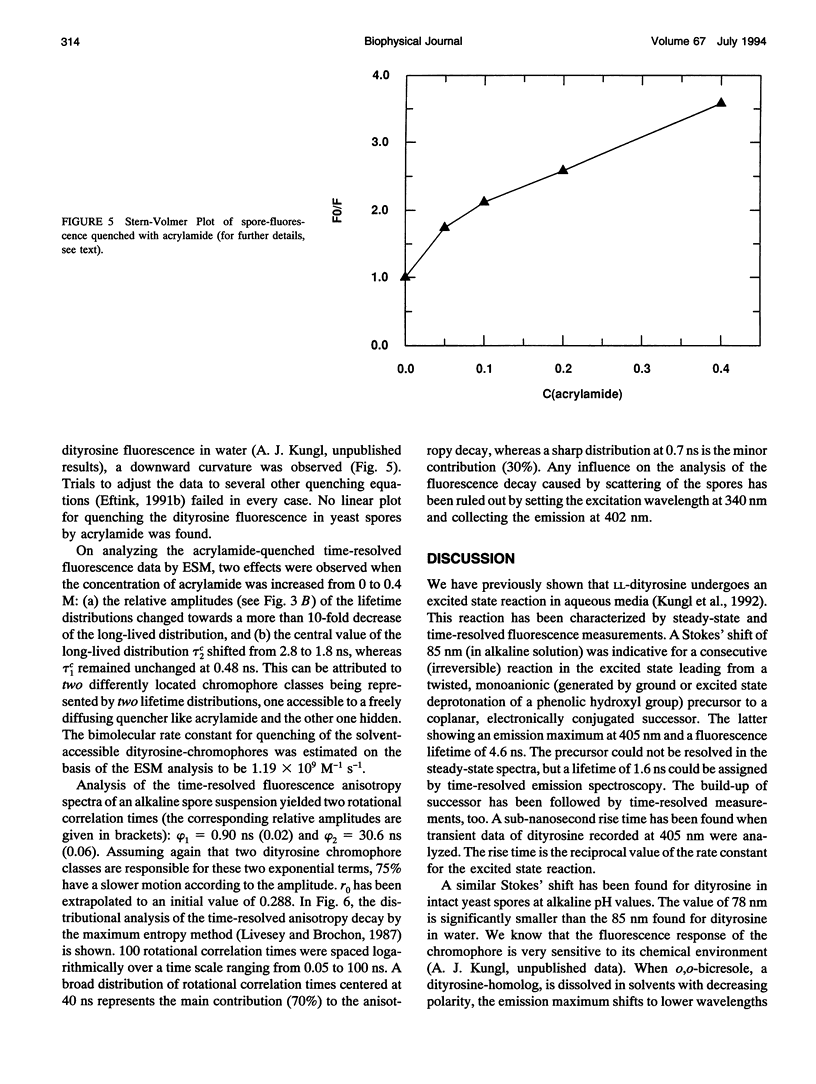
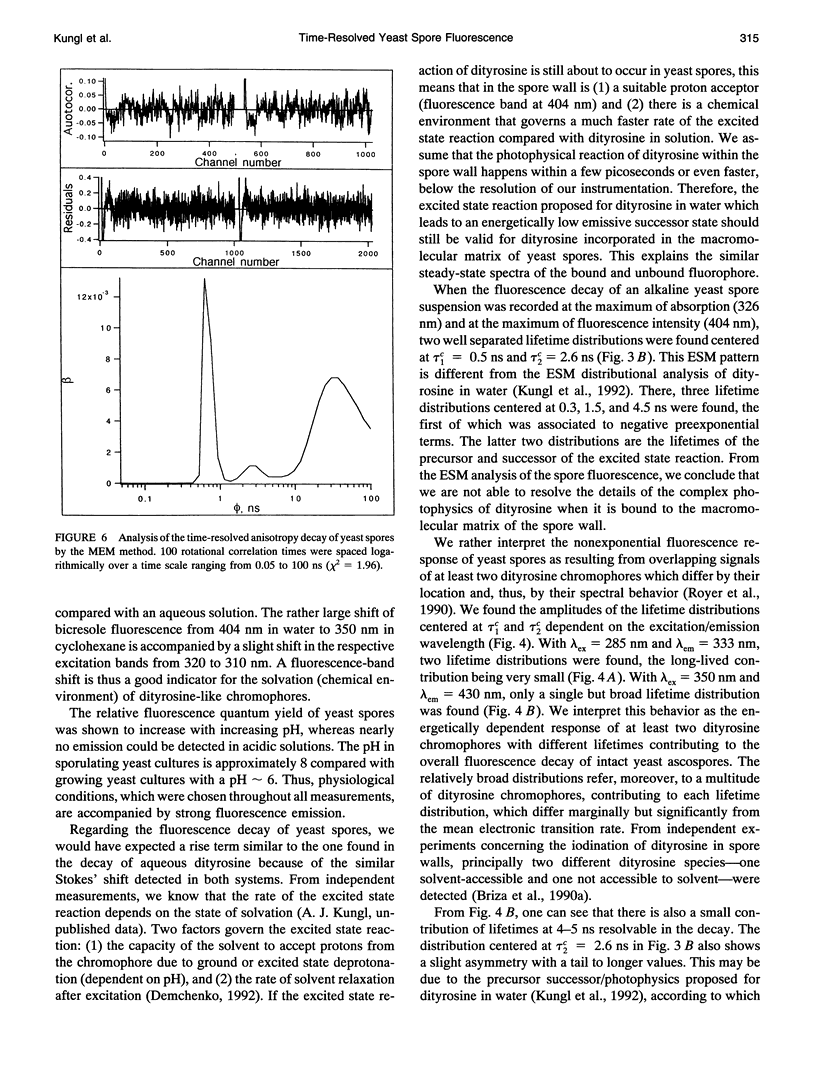
The (time-resolved) fluorescence properties of dityrosine in the outermost layer of the spore wall of Saccharomyces cerevisiae were investigated. Steady-state spectra revealed an emission maximum at 404 nm and a corresponding excitation maximum at 326 nm. The relative fluorescence quantum yield decreased with increasing proton concentration. The fluorescence decay of yeast spores was found to be nonexponential and differed pronouncedly from that of unbound dityrosine in water. Analysis of the spore decay recorded at lambda ex = 323 nm and lambda em = 404 nm by an exponential series (ESM) algorithm revealed a bimodal lifetime distribution with maxima centered at tau 1C = 0.5 ns and tau 2C = 2.6 ns. The relative amplitudes of the two distributions are shown to depend on the emission wavelength, indicating contributions from spectrally different dityrosine chromophores. On quenching the spore fluorescence with acrylamide, a downward curvature of the Stern-Volmer plot was obtained. A multitude of chromophores more or less shielded from solvent in the spore wall is proposed to account for the nonlinear quenching of the total spore fluorescence. Analysis of the fluorescence anisotropy decay revealed two rotational correlation times (phi 1 = 0.9 ns and phi 2 = 30.6 ns) or a bimodal distribution of rotational correlation times (centers at 0.7 ns and 40 ns) when the data were analyzed by the maximum entropy method (MEM). We present a model that accounts for the differences between unbound (aqueous) and bound (incorporated in the spore wall) dityrosine fluorescence. The main feature of the photophysical model for yeast spores is the presence of at least two species of dityrosine chromophores differing in their chemical environments. A hypothetical photobiological role of these fluorophores in the spore wall is discussed: the protection of the spore genome from mutagenic UV light.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alcala J. R., Gratton E., Prendergast F. G. Fluorescence lifetime distributions in proteins. Biophys J. 1987 Apr;51(4):597–604. doi: 10.1016/S0006-3495(87)83384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala J. R., Gratton E., Prendergast F. G. Interpretation of fluorescence decays in proteins using continuous lifetime distributions. Biophys J. 1987 Jun;51(6):925–936. doi: 10.1016/S0006-3495(87)83420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala J. R., Gratton E., Prendergast F. G. Resolvability of fluorescence lifetime distributions using phase fluorometry. Biophys J. 1987 Apr;51(4):587–596. doi: 10.1016/S0006-3495(87)83383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Briza P., Breitenbach M., Ellinger A., Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990 Oct;4(10):1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- Briza P., Ellinger A., Winkler G., Breitenbach M. Characterization of a DL-dityrosine-containing macromolecule from yeast ascospore walls. J Biol Chem. 1990 Sep 5;265(25):15118–15123. [PubMed] [Google Scholar]
- Briza P., Winkler G., Kalchhauser H., Breitenbach M. Dityrosine is a prominent component of the yeast ascospore wall. A proof of its structure. J Biol Chem. 1986 Mar 25;261(9):4288–4294. [PubMed] [Google Scholar]
- Bucci E., Steiner R. F. Anisotropy decay of fluorescence as an experimental approach to protein dynamics. Biophys Chem. 1988 Jul 15;30(3):199–224. doi: 10.1016/0301-4622(88)85017-8. [DOI] [PubMed] [Google Scholar]
- Eftink M. R. Fluorescence techniques for studying protein structure. Methods Biochem Anal. 1991;35:127–205. doi: 10.1002/9780470110560.ch3. [DOI] [PubMed] [Google Scholar]
- Esposito R. E., Dresser M., Breitenbach M. Identifying sporulation genes, visualizing synaptonemal complexes, and large-scale spore and spore wall purification. Methods Enzymol. 1991;194:110–131. doi: 10.1016/0076-6879(91)94010-a. [DOI] [PubMed] [Google Scholar]
- Foerder C. A., Shapiro B. M. Release of ovoperoxidase from sea urchin eggs hardens the fertilization membrane with tyrosine crosslinks. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4214–4218. doi: 10.1073/pnas.74.10.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczynski I., Eftink M., Lakowicz J. R. Conformation heterogeneity in proteins as an origin of heterogeneous fluorescence decays, illustrated by native and denatured ribonuclease T1. Biochim Biophys Acta. 1988 Jun 13;954(3):244–252. doi: 10.1016/0167-4838(88)90079-9. [DOI] [PubMed] [Google Scholar]
- Harris D. L., Hudson B. S. Photophysics of tryptophan in bacteriophage T4 lysozymes. Biochemistry. 1990 Jun 5;29(22):5276–5285. doi: 10.1021/bi00474a009. [DOI] [PubMed] [Google Scholar]
- Hutnik C. M., Szabo A. G. Confirmation that multiexponential fluorescence decay behavior of holoazurin originates from conformational heterogeneity. Biochemistry. 1989 May 2;28(9):3923–3934. doi: 10.1021/bi00435a045. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Maliwal B. P., Cherek H., Balter A. Rotational freedom of tryptophan residues in proteins and peptides. Biochemistry. 1983 Apr 12;22(8):1741–1752. doi: 10.1021/bi00277a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey A. K., Brochon J. C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys J. 1987 Nov;52(5):693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro I., Pecht I., Stryer L. Subnanosecond motions of tryptophan residues in proteins. Proc Natl Acad Sci U S A. 1979 Jan;76(1):56–60. doi: 10.1073/pnas.76.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérola F., Rigler R., Holmgren A., Brochon J. C. Picosecond tryptophan fluorescence of thioredoxin: evidence for discrete species in slow exchange. Biochemistry. 1989 Apr 18;28(8):3383–3398. doi: 10.1021/bi00434a038. [DOI] [PubMed] [Google Scholar]
- Royer C. A., Gardner J. A., Beechem J. M., Brochon J. C., Matthews K. S. Resolution of the fluorescence decay of the two tryptophan residues of lac repressor using single tryptophan mutants. Biophys J. 1990 Aug;58(2):363–378. doi: 10.1016/S0006-3495(90)82383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer C. A. Understanding fluorescence decay in proteins. Biophys J. 1993 Jul;65(1):9–10. doi: 10.1016/S0006-3495(93)81024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Brochon J. C., Merola F., Jordi W., Gallay J. Nanosecond dynamics of horse heart apocytochrome c in aqueous solution as studied by time-resolved fluorescence of the single tryptophan residue (Trp-59). Biochemistry. 1988 Nov 29;27(24):8752–8761. doi: 10.1021/bi00424a010. [DOI] [PubMed] [Google Scholar]
- Vos K., van Hoek A., Visser A. J. Application of a reference convolution method to tryptophan fluorescence in proteins. A refined description of rotational dynamics. Eur J Biochem. 1987 May 15;165(1):55–63. doi: 10.1111/j.1432-1033.1987.tb11193.x. [DOI] [PubMed] [Google Scholar]


