Abstract
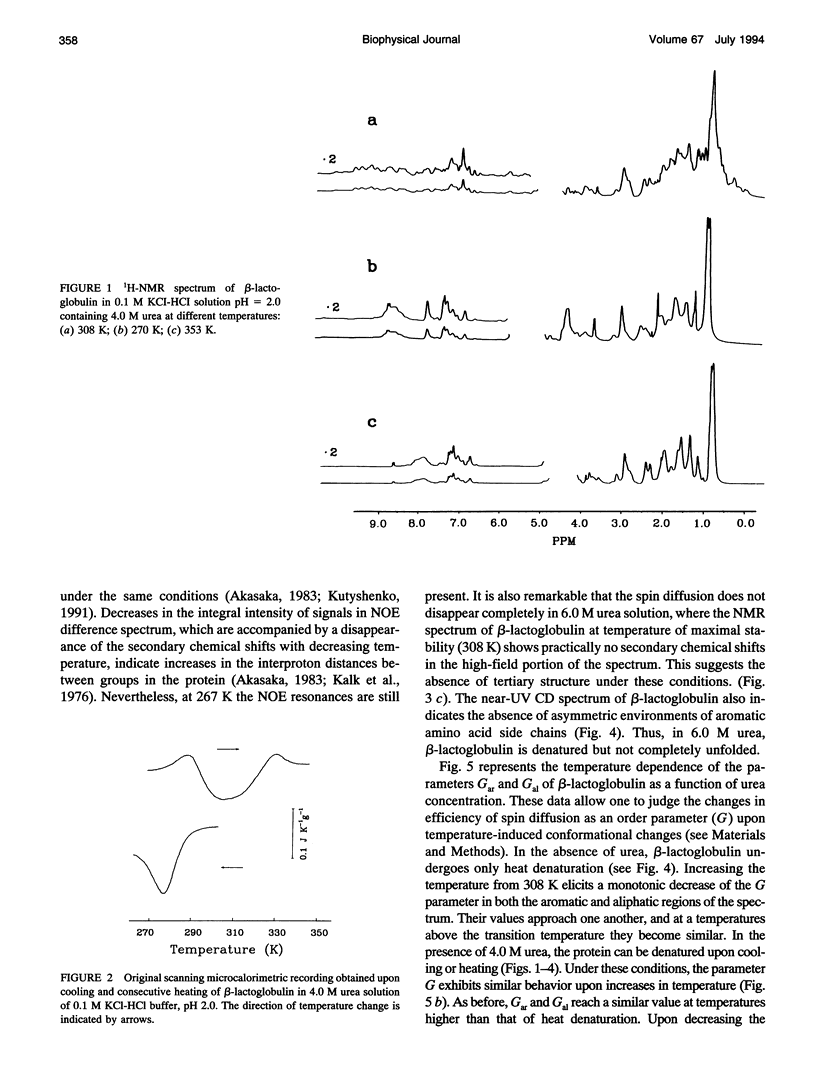
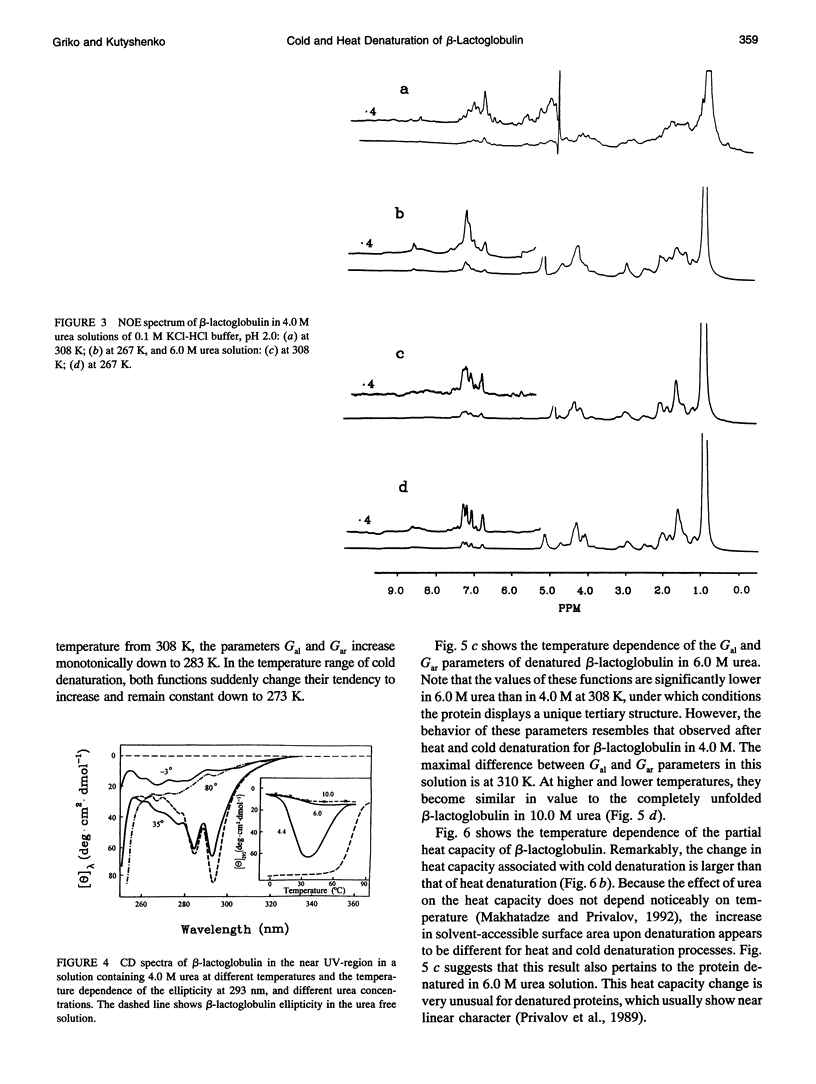
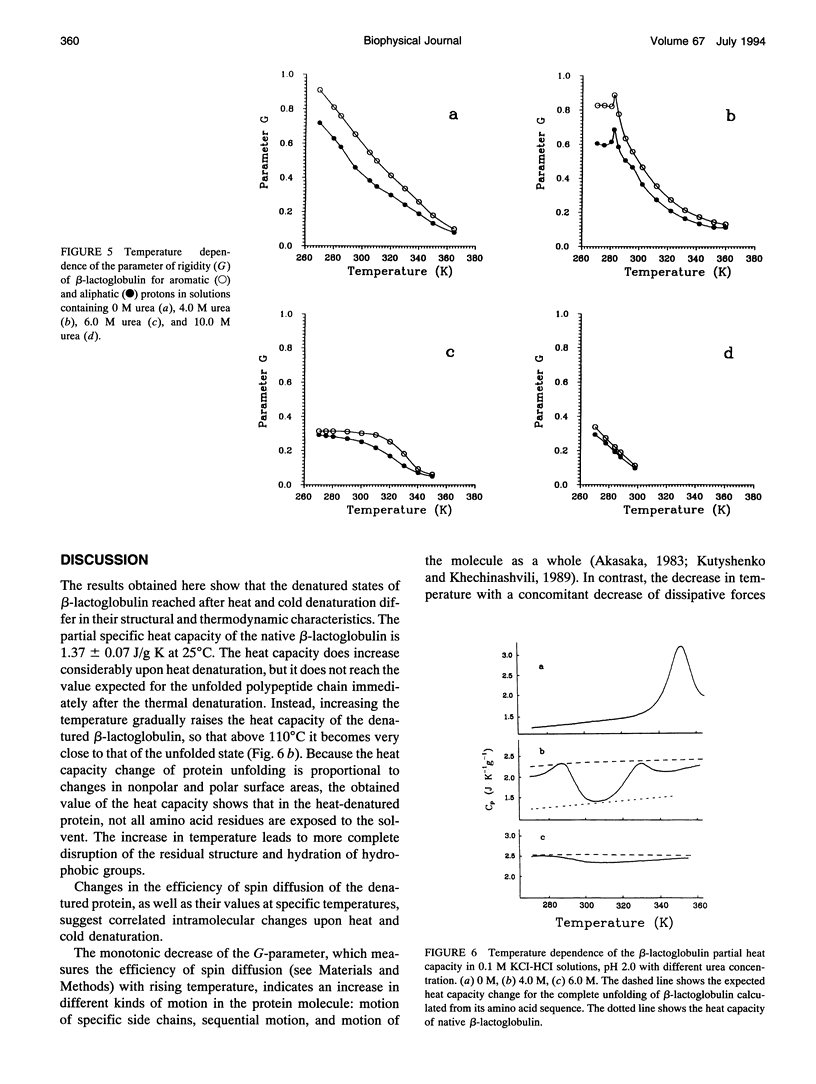
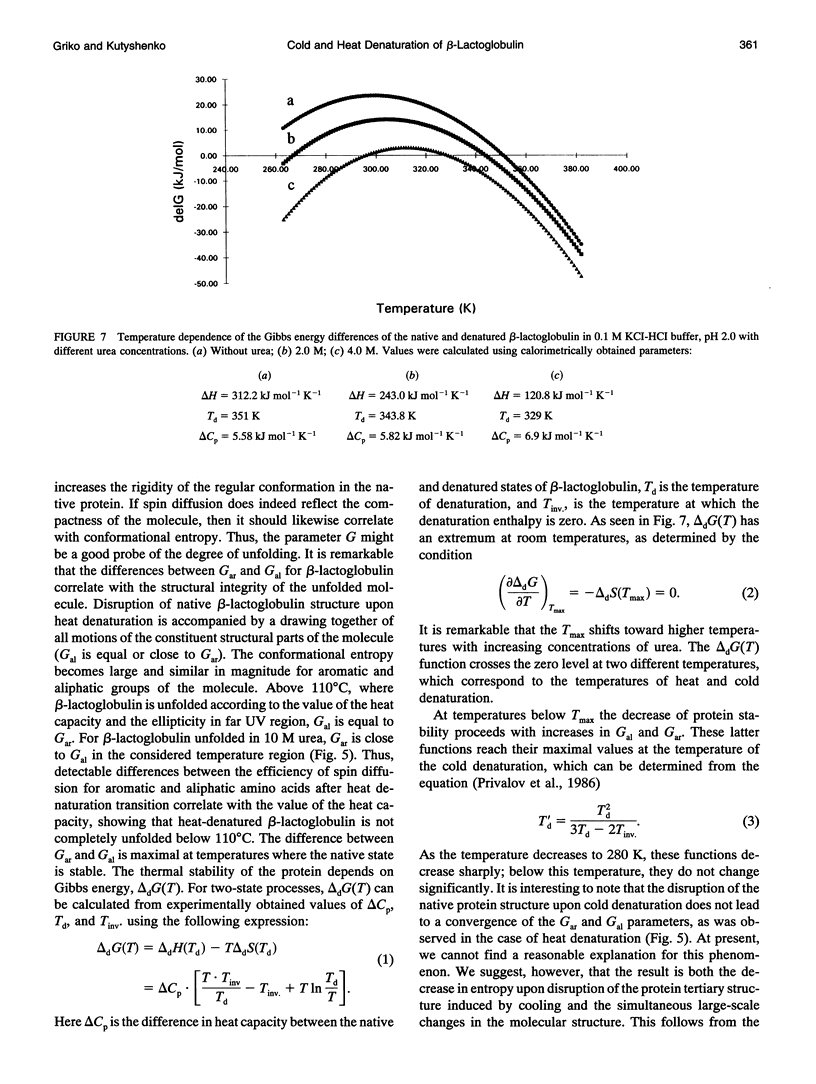
The changes in beta-lactoglobulin upon cold and heat denaturation were studied by scanning calorimetry, CD, and NMR spectroscopy. It is shown that, in the presence of urea, these processes of beta-lactoglobulin denaturation below and above 308 K are accompanied by different structural and thermodynamic changes. Analysis of the NOE spectra of beta-lactoglobulin shows that changes in the spin diffusion of beta-lactoglobulin after disruption of the unique tertiary structure upon cold denaturation are much more substantial than those upon heat denaturation. In cold denatured beta-lactoglobulin, the network of residual interactions in hydrophobic and hydrophilic regions of the molecules is more extensive than after heat denaturation. This suggests that upon cold- and heat-induced unfolding, the molecule undergoes different structural rearrangements, passing through different denaturation intermediates. From this point of view, cold denaturation can be considered to be a two stage process with a stable intermediate. A similar equilibrium intermediate can be obtained at 35 degrees C in 6.0 M urea solution, where the molecule has no tertiary structure. Cooling or heating of the solution from this temperature leads to unfolding of the intermediate. However, these processes differ in cooperativity, showing noncommensurate sigmoidal-like changes in efficiency of spin diffusion, ellipticity at 222 nm, and partial heat capacity. The disruption with cooling is accompanied by cooperative changes in heat capacity, whereas with heating the heat capacity changes only gradually. Considering the sigmoidal shape of the heat capacity change an extended heat absorption peak, we propose that the intermediate state is stabilized by enthalpic interactions.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J. M., McKenzie H. A., Sawyer W. H. On the fractionation of beta-lactoglobulin and alpha-lactalbumin. Biochim Biophys Acta. 1967 Sep 19;147(1):60–72. doi: 10.1016/0005-2795(67)90090-6. [DOI] [PubMed] [Google Scholar]
- Barrick D., Baldwin R. L. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993 Apr 13;32(14):3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- Baum J., Dobson C. M., Evans P. A., Hanley C. Characterization of a partly folded protein by NMR methods: studies on the molten globule state of guinea pig alpha-lactalbumin. Biochemistry. 1989 Jan 10;28(1):7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- Biringer R. G., Fink A. L. Intermediates in the refolding of ribonuclease at subzero temperatures. 3. Multiple folding pathways. Biochemistry. 1988 Jan 12;27(1):315–325. doi: 10.1021/bi00401a048. [DOI] [PubMed] [Google Scholar]
- Biringer R. G., Fink A. L. Observation of intermediates in the folding of ribonuclease A at low temperature using proton nuclear magnetic resonance. Biochemistry. 1982 Sep 14;21(19):4748–4755. doi: 10.1021/bi00262a035. [DOI] [PubMed] [Google Scholar]
- Byler D. M., Susi H., Farrell H. M., Jr Laser-Raman spectra, sulfhydryl groups, and conformation of the cystine linkages of beta-lactoglobulin. Biopolymers. 1983 Dec;22(12):2507–2511. doi: 10.1002/bip.360221204. [DOI] [PubMed] [Google Scholar]
- Casal H. L., Köhler U., Mantsch H. H. Structural and conformational changes of beta-lactoglobulin B: an infrared spectroscopic study of the effect of pH and temperature. Biochim Biophys Acta. 1988 Nov 2;957(1):11–20. doi: 10.1016/0167-4838(88)90152-5. [DOI] [PubMed] [Google Scholar]
- Chan H. S., Dill K. A. Polymer principles in protein structure and stability. Annu Rev Biophys Biophys Chem. 1991;20:447–490. doi: 10.1146/annurev.bb.20.060191.002311. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry. 1990 Aug 7;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Dill K. A., Shortle D. Denatured states of proteins. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- Dobson C. M., Evans P. A., Williamson K. L. Proton NMR studies of denatured lysozyme. FEBS Lett. 1984 Mar 26;168(2):331–334. doi: 10.1016/0014-5793(84)80273-2. [DOI] [PubMed] [Google Scholar]
- Dobson C. M. Protein conformation. Hinge-bending and folding. Nature. 1990 Nov 15;348(6298):198–199. doi: 10.1038/348198a0. [DOI] [PubMed] [Google Scholar]
- Ewbank J. J., Creighton T. E. The molten globule protein conformation probed by disulphide bonds. Nature. 1991 Apr 11;350(6318):518–520. doi: 10.1038/350518a0. [DOI] [PubMed] [Google Scholar]
- Freire E., Biltonen R. L. Thermodynamics of transfer ribonucleic acids: the effect of sodium on the thermal unfolding of yeast tRNAPhe. Biopolymers. 1978 May;17(5):1257–1272. doi: 10.1002/bip.1978.360170512. [DOI] [PubMed] [Google Scholar]
- Freire E., Murphy K. P., Sanchez-Ruiz J. M., Galisteo M. L., Privalov P. L. The molecular basis of cooperativity in protein folding. Thermodynamic dissection of interdomain interactions in phosphoglycerate kinase. Biochemistry. 1992 Jan 14;31(1):250–256. doi: 10.1021/bi00116a034. [DOI] [PubMed] [Google Scholar]
- Griko YuV, Venyaminov SYu, Privalov P. L. Heat and cold denaturation of phosphoglycerate kinase (interaction of domains). FEBS Lett. 1989 Feb 27;244(2):276–278. doi: 10.1016/0014-5793(89)80544-7. [DOI] [PubMed] [Google Scholar]
- Griko Y. V., Privalov P. L. Calorimetric study of the heat and cold denaturation of beta-lactoglobulin. Biochemistry. 1992 Sep 22;31(37):8810–8815. doi: 10.1021/bi00152a017. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Baldwin R. L. Use of site-directed mutagenesis to destabilize native apomyoglobin relative to folding intermediates. Biochemistry. 1989 May 16;28(10):4415–4422. doi: 10.1021/bi00436a044. [DOI] [PubMed] [Google Scholar]
- Hughson F. M., Wright P. E., Baldwin R. L. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990 Sep 28;249(4976):1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- Kim P. S., Baldwin R. L. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lumry R., Biltonen R. Validity of the "two-state" hypothesis for conformational transitions of proteins. Biopolymers. 1966 Sep;4(8):917–944. doi: 10.1002/bip.1966.360040808. [DOI] [PubMed] [Google Scholar]
- Makhatadze G. I., Privalov P. L. Protein interactions with urea and guanidinium chloride. A calorimetric study. J Mol Biol. 1992 Jul 20;226(2):491–505. doi: 10.1016/0022-2836(92)90963-k. [DOI] [PubMed] [Google Scholar]
- Mirsky A. E., Pauling L. On the Structure of Native, Denatured, and Coagulated Proteins. Proc Natl Acad Sci U S A. 1936 Jul;22(7):439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wider G., Wüthrich K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science. 1992 Sep 11;257(5076):1559–1563. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- Neri D., Wider G., Wüthrich K. Complete 15N and 1H NMR assignments for the amino-terminal domain of the phage 434 repressor in the urea-unfolded form. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4397–4401. doi: 10.1073/pnas.89.10.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. C., Tanford C. Thermodynamics of the unfolding of beta-lactoglobulin A in aqueous urea solutions between 5 and 55 degrees. Biochemistry. 1968 Jan;7(1):198–208. doi: 10.1021/bi00841a025. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Griko YuV, Venyaminov SYu, Kutyshenko V. P. Cold denaturation of myoglobin. J Mol Biol. 1986 Aug 5;190(3):487–498. doi: 10.1016/0022-2836(86)90017-3. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Potekhin S. A. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Tiktopulo E. I., Venyaminov SYu, Griko YuV, Makhatadze G. I., Khechinashvili N. N. Heat capacity and conformation of proteins in the denatured state. J Mol Biol. 1989 Feb 20;205(4):737–750. doi: 10.1016/0022-2836(89)90318-5. [DOI] [PubMed] [Google Scholar]


