Abstract
OpcA is an integral outer membrane protein from Neisseria meningitidis, the causative agent of meningococcal meningitis and septicemia. It mediates the adhesion of N. meningitidis to epithelial and endothelial cells by binding to vitronectin and proteoglycan cell-surface receptors. Here, we report the determination of the crystal structure of OpcA to 2.0 Å resolution. OpcA adopts a 10-stranded β-barrel structure with extensive loop regions that protrude above the predicted surface of the membrane. The second external loop adopts an unusual conformation, traversing the axis of the β-barrel and apparently blocking formation of a pore through the membrane. Loops 2, 3, 4, and 5 associate to form one side of a crevice in the external surface of the structure, the other side being formed by loop 1. The crevice is lined by positively charged residues and would form an ideal binding site for proteoglycan polysaccharide. The structure, therefore, suggests a model for how adhesion of this important human pathogen to proteoglycan is mediated at the molecular level.
The colonization of the human host by bacterial pathogens is assisted by specific interactions between microbial surface adhesins and receptors on mammalian cell surfaces. Neisseria meningitidis is a major cause of meningitis and septicemia worldwide (1). It colonizes the nasopharynx asymptomatically and, consequently, adhesion to mucosal cell surfaces is an important part of its life cycle. OpcA is an integral outer membrane protein from N. meningitidis and is functionally similar to the opacity proteins found in Neisseria (2–4). It mediates binding to epithelial and endothelial cells in nonencapsulated strains and functions independently of pilus-based adhesion (5, 6). OpcA attaches to endothelial cells by binding to the serum glycoprotein vitronectin which, in turn, attaches to the integrin αvβ3 (7, 8). OpcA also binds to the same proteoglycan receptors, such as heparin and heparan sulfate, which mediate entry of nonencapsulated meningococci into epithelial cells (9). A homologue of OpcA is present in Neisseria gonorrhoeae: it is expressed at low levels and its function is unclear (10).
Although adhesion to host cells is an important part of the infection process by N. meningitidis, it is poorly understood at the structural level. The crystal structures of several outer membrane proteins have been reported but none were derived from Neisseria species (11). Here, we present the crystal structure of OpcA at 2.0 Å resolution. It reveals that the loop regions from the β-barrel protrude well above the predicted surface of the membrane and form a crevice that would form an ideal site for binding to negatively charged proteoglycan polysaccharide.
Methods
Crystallization and Heavy Atom Derivative Preparation.
Cryo-protected crystals of native and recombinant OpcA were prepared as described (12). Exposure of native OpcA crystals to 100 mM Tris/HCl (pH 7.5)/22% (vol/vol) PEG 6,000/8% (vol/vol) glycerol/0.25% (wt/vol) β-heptyl-glucoside/10 mM CdCl2 for 16 h at 295 K before freezing in liquid nitrogen at 100 K resulted in a drastic change in the unit cell dimensions, from a = 96.9 Å, b = 46.3 Å, and c = 74.0 Å to a = 63.7 Å, b = 50.1 Å, and c = 74.6 Å. We presume that the change is caused by displacement of one or more Zn2+ ions by Cd2+. The CdCl2-treated crystals were in the same space group (P21212) as the standard crystal form, and data from these crystals were subsequently used for multicrystal averaging (see below). All other heavy atom derivatization experiments were conducted in 100 mM Tris/HCl (pH 7.5)/20% (wt/vol) PEG 4,000/200 mM Zn acetate/0.25% (wt/vol) β-heptyl-glucoside/8% glycerol. K2PtCl4 derivatization was conducted at a concentration of 5 mM for 3 h at 295 K. Xe derivatives were prepared by exposing the crystals to 10 bar gauge pressure of Xe gas in an Oxford Cryosystems Xcell for 10 min before flash cooling in liquid nitrogen.
Data Collection.
Data recorded at wavelengths of 0.934 Å and 0.933 Å were collected on beamlines ID14–1 and ID14–2 respectively [European Synchrotron Radiation Facility (ESRF), Grenoble, France]. Data recorded at wavelengths of 0.87 Å and 1.488 Å were collected on beamlines 9.6 and 14.2 respectively [Synchrotron Radiation Source (SRS), Daresbury, U.K.]. All data sets were collected by using 1–3° oscillations on charge-coupled device (CCD) detectors (ADSC Quantum4 for SRS beamlines, MAR CCD at the ESRF beamlines) and processed with the program MOSFLM (13). Further computations were carried out by using programs from the Collaborative Computational Project Number 4 (CCP4) suite (14) except where stated otherwise.
Phasing.
An initial phase set was obtained from xenon and platinum derivatives of recombinant OpcA crystals (Table 1). A single Xe site was identified from the anomalous difference Patterson map calculated from the Xe derivative data collected at 1.488 Å wavelength; the position of the Xe atom was solved by inspection. Single isomorphous replacement (SIR) phases from this derivative suffered from P222 pseudosymmetry because of the location of this Xe site (fractional coordinates x, y, z at 0.25, 0.25, 0.03, respectively). The heavy atom substructure of the K2PtCl4 derivative was solved by crossphasing the anomalous difference amplitudes from the data set recorded at 0.87 Å. The pseudosymmetry was broken by the incremental inclusion of Pt sites while checking against the combined K2PtCl4 and Xe derivative. Subsequent multiple isomorphous replacement with anomalous scattering (MIRAS) crossphasing located an additional two Xe minor sites. MIRAS phases were calculated from all derivative data sets by using the program SHARP (15), and an initial solvent flattened map was prepared by using the program DM (16).
Table 1.
Data collection, phasing and refinement statistics
| Data set | Native
|
Recombinant
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| CdCl2 | Xe | K2PtCl4 | K2PtCl4 + Xe | ||||||
| Wavelength, Å | 0.933 | 0.934 | 1.488 | 0.87 | 1.488 | 0.87 | 1.488 | 0.87 | 0.87 |
| Resolution, Å | 36.4–2.0 | 34.8–2.7 | 48.8–2.8 | 41.3–2.6 | 58.7–2.8 | 40.8–2.5 | 36.3–2.8 | 58.7–3.0 | 25.0–2.8 |
| Completeness, % | 98.3 | 99.6 | 99.6 | 98.9 | 100 | 97.3 | 99.5 | 99.9 | 99.7 |
| I/σ(I) | 16.5 | 12.8 | 25.6 | 11.0 | 38.6 | 11.9 | 16.9 | 30.0 | 18.1 |
| Redundancy | 10.2 | 5.7 | 6.3 | 3.4 | 15.1 | 3.3 | 4.9 | 16.2 | 6.5 |
| Rmerge, %* | 8.1 | 7.4 | 5.1 | 6.7 | 5.7 | 5.2 | 5.5 | 7.7 | 5.5 |
| Riso, %† | – | – | – | – | 22.8 | 23.0 | 24.0 | 23.0 | 27.9 |
| Acentric Rcullis‡ | – | – | – | – | 0.80 | 0.81 | 0.94 | 0.94 | 0.95 |
| Anom. Rcullis‡ | 0.70 | 0.89 | 0.91 | 0.84 | 0.90 | ||||
| Centric Rcullis‡ | – | – | – | – | 0.85 | 0.86 | 0.90 | 0.91 | 0.92 |
| No. sites | – | – | – | – | 3 | 3 | 7 | 7 | 7 + 3 |
Rmerge = ∑hkl∑sym|I − 〈I〉|/∑hklI;
Riso = ∑hkl|Fder − Fnat|/∑hklFnat
Isomorphous Rcullis = ∑hkl∥Fphobs| − |Fphcalc∥/∑hkl|Fph − Fp| = 〈loc〉/〈iso.diff.〉; (anomalous Rcullis = ∑hkl∥F+ − F−| − (2x|Fh"calc|)|/∑hkl|F+ − F−| = 〈loc〉/〈ano.diff.〉)
Molecular Replacement Solution of the CdCl2 Crystal Form.
A polyalanine chain representing approximately 75% of the OpcA polypeptide was built by using the program XFIT (17). Phased refinement and phase combination with the program REFMAC (18) led to an improved map from which the OpcA sequence was assigned to the polyalanine trace. Three spherically symmetric ion sites were located in the electron-density maps; all appeared to be tetrahedrally coordinated and were interpreted as Zn2+ ions. This partially refined model (R, 42.8%; Rfree, 45.1%) was used to obtain a molecular replacement solution for the CdCl2 dataset over the resolution interval 16–4 Å with the program AMORE (ref. 19; solution: Euler angles αβγ −23.9, 4.8, 221.06; fractional translations 0.239, 0.152, 0.089; correlation coefficient, 65.6%; R-factor, 44.2%). This solution, along with a mask generated from the model, was used with the CdCl2 dataset to transfer the MIRAS rOpcA phase set to the native OpcA 2.0 Å data by a process of multicrystal averaging with the program DMM (16). The OpcA model was rebuilt into this improved, unbiased, electron density.
Model Building and Refinement.
Initially, 10 rigid bodies comprising periplasmic and extra-membranous facing loops were defined, and translation, libration, and screw rotation (TLS) tensors were refined for each (20). This procedure allowed the incremental building of the disordered outer membrane loops by using the program O (21). Finally, the TLS rigid-body set was extended to 28 groups, and residual isotropic B-factors were refined. The final model contained 248 residues in a single polypeptide chain (N-terminal residues 1–4 were not seen in the maps), 3 Zn ions, 144 water molecules, and the head groups of 2 C10E5 detergent molecules (Table 2). Equivalent B-factors were calculated from TLS tensors and residual B-factors after refinement in REFMAC (18). The stereo-chemical quality of the final protein model was better than that expected for a 2.0 Å model, as judged by the program PROCHECK (22).
Table 2.
Refinement statistics
| Data set | |
|---|---|
| No. of reflections | 20,754 (2,561)* |
| rms deviation bond lengths, Å | 0.019 |
| rms deviation angles, ° | 1.951 |
| Rcryst, % | 23.5 (22.7) |
| R-free (5% of reflections) | 27.4 (28.8) |
| Estimated coordinate error, Å (based on R-free) | 0.181† |
| Total number of refined atoms | 2,131 |
| Number of water molecules | 144 |
Figures in parentheses denote values for the outer resolution bin (9.1% of refinement data).
As implemented in refmac (18).
Results
Overall Structure.
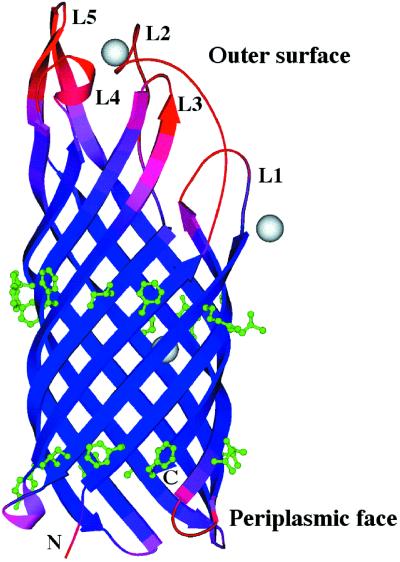
The overall shape of the OpcA monomer is a uniform flattened cylinder, measuring approximately 65 Å in length and consisting of a 10-standed transmembrane β-barrel with a shear number of 12 (ref. 23; Fig. 1). All outer-membrane protein crystal structures determined to date consist of a βbarrel fold, and structures with 8–18 and 22 β-strands have now been determined (11, 24, 25). The majority of these proteins were derived from Escherichia coli. Although crystals of recombinant OpcA were used for obtaining derivatives, the highest resolution data were obtained from native OpcA isolated without denaturation directly from N. meningitidis. The cross section of the β-barrel is uniformly flattened along its length, with maximal and minimal diameters of 23 and 15 Å respectively. The interior of the protein is packed with side chains from hydrophilic amino acids that are hydrogen bonded to a network of water molecules. The membrane-spanning portion of the barrel was readily identified from the hydrophobic side chains that protrude outwards (Fig. 2). This ‘inverse micelle’ has been observed in other outer-membrane protein structures (11). There are two rings of aromatic residues flanking the top and bottom of the barrel, similar to other integral membrane proteins (Fig. 1). One end of the barrel consists of much shorter turns and forms the periplasmic end of the molecule, whereas the exterior is made up of five loop regions of varying length that would protrude above the membrane. These assignments are similar to a two-dimensional topology map of OpcA proposed previously, except that the location of the periplasmic turns within the sequence differ by up to four amino acids (26). The loops correspond to regions that are known to be exposed on the cell surface from antibody reactivity against an inserted antigen sequence (26). The β-strands extend well above the predicted membrane surface, with this regular secondary structure contributing to the formation of the loop regions. The loops are more mobile than the membrane-spanning β-strands, as indicated by their disorder (Fig. 1).
Figure 1.
Structure of N. meningitidis OpcA. The chain is color-coded for equivalent B-factors, with lower values in blue to higher values in red. Three Zn2+ ions are shown in gray, and the two rings of hydrophobic residues are shown in green. The figure was generated by using MOLSCRIPT (42).
Figure 2.
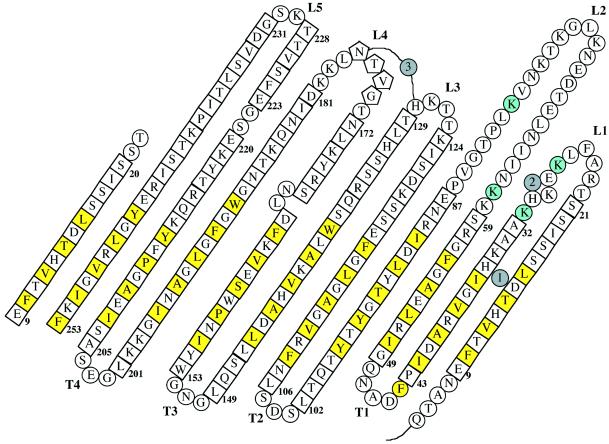
Topology diagram of OpcA as viewed from the outside of the barrel. Residues in a β-strand are shown in squares, those in coil are shown in circles, and those in helix are shown in pentagons. The locations of the three Zn2+ ions are shown in gray circles. Hydrophobic residues, which extend outwards toward the membrane, are highlighted in yellow. Lys-27, -31, -61, and -80, which form part of the proposed binding site for heparin, are highlighted in cyan. The first strand is shown twice to show the interaction between the first and last strands in the barrel.
Comparison with OmpT Protease.
A comparison of the structure of OpcA with the E. coli OmpT protease, the only other known 10-stranded β-barrel OMP (25), showed that the backbone atoms from the transmembrane β-strands overlaid with an rms deviation of 1.6 Å. Although the overall fold of the two proteins was similar, three important differences were noted. First, the N and C termini were located in different positions, with the first β-strand in OpcA aligning with the seventh β-strand in OmpT. Second, the exterior loop regions in OmpT are flared outwards from the central axis of the barrel, whereas the equivalent loops in OpcA remain continuous with the transmembrane β-barrel. Third, the overall shape of the OpcA β-barrel was more uniform than OmpT: a length-wise cross section of the OmpT barrel shows that it narrows significantly in the central portion of the barrel. In contrast, the OpcA barrel is much more uniform along its length. It is possible that this difference reflects two contrasting solutions to the problem of blocking pore formation. Both barrels are apparently accessible to water from the periplasmic side, but the channel in OpcA is effectively blocked by the longest loop, L2. There is a pronounced kink in L2 at Glu-87, which causes it to traverse the central axis of the barrel before extending outwards. In the case of OmpT, the channel is occluded by the direct interaction of side chains from within the barrel itself.
Crystal Packing and Oligomerization.
Both the crystal forms of OpcA (standard and CdCl2-treated) are type I membrane protein crystals and contain one protein monomer in the asymmetric unit. None of the crystal packing interactions suggest that OpcA forms multimers in the detergent-solubilized state. This conclusion does not preclude its function as a proteoglycan-binding protein but is in apparent contradiction to previous evidence that OpcA is a trimer in zwittergent micelles (2) and when reconstituted into dimyristoyl phosphatidyl choline lipid bilayers (27). It is possible that these monomer–monomer interactions are relatively weak and are disrupted by the stronger ionic strength conditions used for crystallization. The predominant crystal packing interactions are between the apolar side chains on the surface of the β-barrel, forming the main monomer–monomer interaction along a 21 symmetry axis. The presence of Zn2+ ions was mandatory for crystal formation (12), and three separate peaks of high electron density were attributed to Zn2+. The crystal packing interactions between the more mobile loop regions are mediated by Zn2+, indicating that they play an important part in crystal formation by generating specific contacts between OpcA molecules. The third ion is located within the central cavity in the interior of the β-barrel and is coordinated by His-13, Asp-15, His-35, and a water molecule. No Zn2+-containing buffers were included in the purification media, so this ion was possibly introduced during the crystallization process. Two patches of elongated electron density were observed adjacent to turns T1 and T3 on the periplasmic side of the molecule; these were built as head groups of the detergent C10E5, which was used for crystallization (12).
Loop Structure and Crevice Formation.
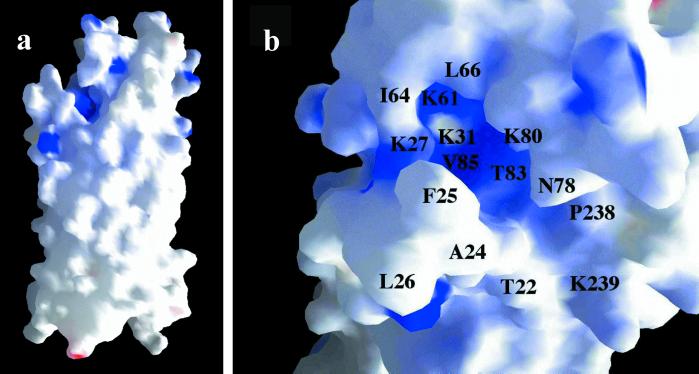
The five loop regions associate to form a continuous surface on the external face of OpcA. Loops L3, L4, and L5 are located directly over the sides of the barrel in a similar fashion to the loop regions in the 8-stranded E. coli OmpA and OmpX outer-membrane proteins (28, 29). L2 is located in a central position at the top of the barrel and interacts with L3, L4, and L5 to form a continuous surface. This interaction involves burying apolar residues; for example, Leu-81 from loop L2 forms a well defined hydrophobic pocket with Phe-224, Val-226, Leu-171, Leu-178, Ile-182, and Leu-235. L1, however, is separate from the other loop regions and does not make any direct interaction with them. This effectively generates a crevice ≈8 Å wide, 22 Å long, and 11 Å deep in the external surface of OpcA, which is lined by residues from L1, L2, and L5 (Fig. 3a). Examination of the electrostatic potential across the surface showed that the crevice contained a high proportion of basic residues, principally Lys-27, -31, -61, and -80 (Fig. 3b). We propose that this crevice is the location of the heparin/proteoglycan-binding site for the following reasons. First, we would anticipate that the binding site is located on the outer surface of the protein; the crevice is the dominant feature in the surface structure of the external loop regions. Second, structural studies of heparin-binding sites in other proteins have shown a preponderance of basic side chains (30–35). Third, the dimensions of the crevice could plausibly accommodate a heparin polysaccharide (36).
Figure 3.
Surface representations of OpcA, colored by electrostatic potential with blue as positive charge and red as negative charge. (a) Structure of a whole OpcA monomer. The proposed proteoglycan-binding site with its concentration of positive charge is visible at the top left of the molecule. (b) Detail of the proposed binding site, viewed from above, with specific residues marked. The diagram was generated by using GRASP (43).
The location of the proposed binding site, 15–20 Å from the membrane-defining upper annulus of aromatic residues, provides a rationale for the continuation of the β-barrel well above the membrane surface: this is required to facilitate access to proteoglycan chains. A similar phenomenon is observed in OmpX, where four β-strands are arranged to form a protruding sheet motif which has been postulated to have a role in mediating adhesion to unidentified receptors (29). The arrangement of the loop regions in OpcA appears to be designed to form a more complex binding surface. The loops also contain a number of exposed hydrophobic residues which could play a role in recognition of polysaccharide, for example, Phe-25 or Leu-26. It seems plausible, judging from the higher mobility of the loop regions, that there could be some form of rearrangement of the loop residues on binding of proteoglycan.
Discussion
The structural basis for heparin recognition by proteins seems to vary widely but is characterized by a strong preference for basic residues within the binding site. In the case of the NK1-heparin complex, for example, the binding site consists of a shallow crevice, formed by the edge of an adjacent β-strand, a helix, and a connecting loop (30). The binding site for the heparin decasaccharide in the complex with the fibroblast growth factor and its receptor is more complicated; it is distributed between two growth factor molecules and a receptor (31). None of the heparin-binding sites in this structure forms a well defined crevice, but they all contain Lys and Arg residues which form specific hydrogen bonds to sulfate groups in the heparin ligand. In other cases, structures of heparin-binding proteins are available in the unliganded state, and supporting biochemical information has helped to identify the heparin-binding site (33–35). The proposed heparin-binding site on OpcA is notable for the pronounced crevice structure and also for the patch of positive charge in this region. In this respect at least, the structure of OpcA appears to have some features in common with other heparin-binding proteins.
The only known homologue of OpcA from N. meningitidis is a protein (also termed OpcA) from the related bacterium N. gonorrhoeae (10). A sequence alignment indicated a high degree of sequence conservation in the transmembrane β-strand regions but more variability in the loops. There is a deletion in that part of loop L2 which is responsible for plugging the β-barrel (Pro-82 to Pro-86), although Glu-87 is conserved. All of the four lysines within the proposed heparin-binding site (Lys-27, -31, -61, and -80) are conserved in the alignment given by Zhu et al. (10). It should be noted, however, that much of this interpretation depends on the correct alignment of the sequences within the loop L2, which is unclear because of the sequence diversity in this region. Also, it is important to emphasize that Zhu et al. could only find evidence for low-level transcription and expression of the OpcA gene in N. gonorrhoeae, and there is no direct evidence that this protein is able to bind to heparin.
From a more general perspective, there is now a considerable body of evidence that cell-surface proteoglycans act as receptors for bacterial adhesins (reviewed in ref. 37). Organisms as diverse as Listeria (38), Helicobacter pylori (39), and Pseudomonas aeruginosa (40) have been shown to bind to heparin and heparan sulfate. In the case of Neisseria, adhesion has been shown to be mediated by OpcA in N. meningitidis (9) and the OpaA protein in N. gonorrhoeae (41). The interaction of N. meningitidis with epithelial cells has been proposed to occur in a two-stage process, with an initial localized adhesion step mediated by pili and a second diffuse adherence step, perhaps involving proteins from the OpcA/Opa protein families (4). The current knowledge of the three-dimensional structures of bacterial adhesins is limited, particularly for those that are intrinsic membrane proteins. The crystal structure of OpcA has provided some important clues as to how outer-membrane protein structures could be adapted to function as adhesins. These observations suggest an analogy with antibody-binding sites, which also consist of loop regions built onto a β-structure framework. Such a design provides a great deal of flexibility and indicates how outer membrane proteins could be modified to bind to a wide range of ligands on the surface of host cells. It also raises the possibility that drugs could be designed, by using structural information, to inhibit adhesion of meningococci to the human host.
Acknowledgments
We thank the staff at European Synchrotron Radiation Facility, Grenoble, France and Synchrotron Radiation Source Daresbury, U.K. for assistance with data collection. We thank Dr. P. Bullough for valuable discussions. This work was funded by Grant 056289 from the Wellcome Trust. We gratefully acknowledge support from the North of England Structural Biology Consortium, funded by the Biotechnology and Biological Sciences Research Council. J.P.D. is a Fellow of the Lister Institute of Preventive Medicine.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1K24).
References
- 1.Cartwright K A V. Meningococcal Disease. Chichester, U.K.: John Wiley & Sons; 1995. [Google Scholar]
- 2.Achtman M, Neibert M, Crowe B A, Strittmatter W, Kusecek B, Weyse E, Walsh M J, Slawig B, Morelli G, Moll A, Blake M. J Exp Med. 1988;168:507–525. doi: 10.1084/jem.168.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malorny B, Morelli G, Kusecek B, Kolberg J, Achtman M. J Bacteriol. 1998;180:1323–1330. doi: 10.1128/jb.180.5.1323-1330.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nassif X, Pujol C, Morand P, Eugene E. Mol Microbiol. 1999;32:1124–1132. doi: 10.1046/j.1365-2958.1999.01416.x. [DOI] [PubMed] [Google Scholar]
- 5.Virji M, Makepeace K, Ferguson D J P, Achtman M, Moxon E R. Mol Microbiol. 1993;10:499–510. doi: 10.1111/j.1365-2958.1993.tb00922.x. [DOI] [PubMed] [Google Scholar]
- 6.Virji M, Makepeace K, Ferguson D J P, Achtman M, Sarkari J, Moxon E R. Mol Microbiol. 1992;6:2785–2795. doi: 10.1111/j.1365-2958.1992.tb01458.x. [DOI] [PubMed] [Google Scholar]
- 7.Virji M, Makepeace K, Peak I R A, Ferguson D J P, Jennings M P, Moxon E R. Mol Microbiol. 1995;18:741–754. doi: 10.1111/j.1365-2958.1995.mmi_18040741.x. [DOI] [PubMed] [Google Scholar]
- 8.Virji M, Makepeace K, Moxon E R. Mol Microbiol. 1994;14:173–184. doi: 10.1111/j.1365-2958.1994.tb01277.x. [DOI] [PubMed] [Google Scholar]
- 9.de Vries F P, Cole R, Dankert J, Frosch M, van Putten J P M. Mol Microbiol. 1998;27:1203–1212. doi: 10.1046/j.1365-2958.1998.00763.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhu P X, Morelli G, Achtman M. Mol Microbiol. 1999;33:635–650. doi: 10.1046/j.1365-2958.1999.01514.x. [DOI] [PubMed] [Google Scholar]
- 11.Koebnik R, Locher K P, van Gelder P. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 12.Prince S M, Feron C, Janssens D, Lobet Y, Achtman M, Kusecek B, Bullough P A, Derrick J P. Acta Crystallogr D. 2001;57:1164–1166. doi: 10.1107/s0907444901009416. [DOI] [PubMed] [Google Scholar]
- 13.Leslie A G W. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1992. 26. [Google Scholar]
- 14.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 15.de la Fortelle E, Bricognne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 16.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 17.McRee D E. J Mol Graph. 1992;10:44–46. [Google Scholar]
- 18.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 19.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 20.Winn M D, Isupov M N, Murshudov G N. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 21.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–293. [Google Scholar]
- 23.Murzin A G, Lesk A M, Chothia C. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 24.Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Nature (London) 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 25.Vandeputte-Rutten L, Kramer R A, Kroon J, Dekker N, Egmond M R, Gros P. EMBO J. 2001;20:5033–5039. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merker P, Tommassen J, Kusecek B, Virji M, Sesardic D, Achtman M. Mol Microbiol. 1997;23:281–293. doi: 10.1046/j.1365-2958.1997.2051567.x. [DOI] [PubMed] [Google Scholar]
- 27.Collins R, Achtman M, Ford R, Bullough P, Derrick J. Mol Microbiol. 1999;32:217–219. doi: 10.1046/j.1365-2958.1999.01335.x. [DOI] [PubMed] [Google Scholar]
- 28.Pautsch A, Schultz G E. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 29.Vogt J, Schultz G E. Structure (London) 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 30.Lietha D, Chirgadze D Y, Mulloy B, Blundell T L, Gherardi E. EMBO J. 2001;20:5543–5555. doi: 10.1093/emboj/20.20.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellegrini L, Burke D F, von Delft F, Mulloy B, Blundell T L. Nature (London) 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 32.DiGabriele A D, Lax I, Chen D I, Svahn C M, Jaye M, Schlessinger J, Hendrickson W A. Nature (London) 1998;393:812–817. doi: 10.1038/31741. [DOI] [PubMed] [Google Scholar]
- 33.Tisi D, Talts J F, Timpl R, Hohenester E. EMBO J. 2000;19:1432–1440. doi: 10.1093/emboj/19.7.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong J, Peters-Libeu C A, Weisgraber K H, Segelke B W, Rupp B, Capila I, Hernaiz M J, LeBrun L A, Linhardt R J. Biochemistry. 2001;40:2826–2834. doi: 10.1021/bi002417n. [DOI] [PubMed] [Google Scholar]
- 35.Skinner R, Abrahams J P, Whisstock J C, Lesk A M, Carrell R W, Wardell M R. J Mol Biol. 1997;266:601–609. doi: 10.1006/jmbi.1996.0798. [DOI] [PubMed] [Google Scholar]
- 36.Mulloy B, Forster M J, Jones C, Davies D B. Biochem J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rostand K S, Esko J D. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Dominguez C, Vazquez-Boland J A, Carrasco-Marin E, Lopez-Mato P, Leyva-Cobian F. Infect Immun. 1997;65:78–88. doi: 10.1128/iai.65.1.78-88.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guzman-Murillo M A, Ruiz-Bustos E, Ho B, Ascencio F. J Med Microbiol. 2001;50:320–329. doi: 10.1099/0022-1317-50-4-320. [DOI] [PubMed] [Google Scholar]
- 40.Plotkowski M C, Costa A O, Morandi V, Barbosa H S, Nader H B, De Bentzmann S, Puchelle E. J Med Microbiol. 2001;50:183–190. doi: 10.1099/0022-1317-50-2-183. [DOI] [PubMed] [Google Scholar]
- 41.van Putten J P M, Paul S M. EMBO J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 43.Nicholls A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]