Abstract
The SsrA or tmRNA quality control system intervenes when ribosomes stall on mRNAs and directs the addition of a C-terminal peptide tag that targets the modified polypeptide for degradation. Although hundreds of SsrA-tagged proteins can be detected in cells when degradation is prevented, most of these species have not been identified. Consequently, the mRNA sequence determinants that cause ribosome stalling and SsrA tagging are poorly understood. SsrA tagging of Escherichia coli ribokinase occurs at three specific sites at or near the C terminus of this protein. The sites of tagging correspond to ribosome stalling at the termination codon and at rare AGG codons encoding Arg-307 and Arg-309, the antepenultimate and C-terminal residues of E. coli ribokinase. Mutational analyses and studies of the effects of overexpressing the tRNA that decodes AGG reveal that the combination of a rare arginine codon at the C terminus and the adjacent inefficient UGA termination codon act to recruit the SsrA-tagging system, presumably by slowing the rate of translation elongation and termination.
All eubacteria possess a quality-control system, the SsrA- or tmRNA-tagging system, which frees ribosomes stalled at the 3′ ends of truncated mRNAs and directs the proteolysis of proteins synthesized from these messages (1–3). SsrA from Escherichia coli is an RNA molecule that has been shown to function both as a transfer RNA and a messenger RNA (1–3). The 5′ and 3′ ends of SsrA fold into a tRNA-like domain that is aminoacylated with alanine (4, 5). SsrA also contains a short ORF, which in E. coli encodes a decapeptide sequence (ANDENYALAA) (1, 6). According to the tmRNA model, alanylated SsrA binds to the vacant A-site on a stalled ribosome, and the charged alanine is added to the nascent polypeptide by transpeptidation (1, 2). The original mRNA is then released by the ribosome and translation resumes by using the short ORF within SsrA, resulting in a product with the SsrA-derived peptide tag at the C terminus of the truncated protein. SsrA-tagged proteins are then degraded by intracellular proteases (1, 7, 8).
SsrA tagging has been demonstrated by using artificially truncated mRNAs lacking in-frame stop codons (1). However, SsrA tagging can also be directed by full-length messages, either at genetically engineered clusters of rare codons or at termination codons, and proteomic studies show that several hundred different proteins are tagged at low levels by the SsrA system (9–11). These observations suggest that SsrA activity may play a regulatory role in gene expression. Indeed, good evidence suggests that the SsrA-tagging system modulates levels of the LacI repressor, and the activities of several transcription factors that are important for phage gene expression and development (10–13).
Although proteins encoded by natural full-length mRNAs can be tagged by the SsrA system (9), no detailed studies have been made of the sites of tagging nor of the mRNA determinants that cause tagging. The E. coli enzyme ribokinase (RbsK) is one such example (9). Ribokinase catalyzes the conversion of ribose to ribose 5-phosphate; its gene, rbsK, is part of the rbs operon, which also encodes high-affinity ribose transport proteins (14, 15). Here, we have identified the primary sites of SsrA tagging in RbsK and analyzed how mutations in rbsK alter tagging at these sites. These studies show that an inefficient translation termination codon (UGA) and the adjacent rare codon encoding Arg-309 (AGG) cause 10–25% of newly synthesized RbsK molecules to be tagged by the SsrA system. Rare arginine codons adjacent to stop codons are found in E. coli genes at levels that exceed the statistically expected frequency and apparently are a general determinant of SsrA tagging.
Experimental Procedures
Bacterial Strains and Plasmids.
E. coli strains were derivatives of X90 [F′ lacIq lac′ pro′/ara Δ(lac-pro) nalA argE(am) rifr thi-1]. Strain X90 ssrA∷cat has been described (9). Strain X90 ssrA∷cat (DE3) was generated by using the DE3 lysogenization kit from Novagen. Plasmids pRbsK1 and pRbsK2 express ribokinase from wild-type rbsK genes, under control of a Ptrc promoter which is inducible by isopropyl β-D-thiogalactoside (IPTG). These plasmids were constructed by PCR amplification of the rbsK gene from E. coli X90 genomic DNA by using the following oligonucleotide primers that contain restriction endonuclease sites (underlined residues) to facilitate cloning: RBSK-FOR, 5′-GGT GGC GCA TTC CAT GGA CAT CCC G (upstream primer); RBSK-REV1, 5′-GTT CTT GGA TCC CCG CTT CAA CTT TGG (downstream primer 1); and RBSK-REV2, 5′-CAT TGT GGA TCC GCG TCA CCT CTG CCT GTC TAA (downstream primer 2). The resulting products were digested with NcoI and BamHI and ligated to identically digested plasmid pTrc99a (Amersham Pharmacia). Plasmid pRbsK1 contains the entire rbsK coding region plus 115 bp of downstream sequence containing the 5′ end of the rbsR gene. Plasmid pRbsK2 contains only 3 bp of sequence downstream of the rbsK stop codon. All site-directed mutant forms of the rbsK gene were generated by using the QuikChange (Stratagene) protocol with plasmid pRbsK1 as template DNA.
Plasmids pRpiB-RRN, pRpiB-RQR, and pYjgR are expression plasmids that use the T7 RNA polymerase promoter. pRpiB-RRN expresses wild-type ribose-5-phosphate isomerase (RpiB) from E. coli, and plasmid pRpiB-RQR expresses a mutant version of RpiB in which the last three codons and the stop codon of rpiB have been replaced with the AGG CAG AGG TGAC nucleotide sequence from the 3′ end of the rbsK gene. These plasmids were constructed with PCR-amplified fragments by using the RPIB-FOR forward primer (5′-AAG ATC CAT GGC TAA AAA GAT TGC ATT TGG CTG TG), with primer RPIB-RRN (5′-GGA TCC ATC TCA ATT TCT CCG CTG C) for the wild-type rpiB gene and primer RPIB-RQR (5′-GGA TCC ATG TCA CCT CTG CCT CTG CTC TAT TGC CGT AAT CGC CTC C) for the mutant rpiB gene. The rpiB gene fragments were digested with NcoI and BamHI and ligated to NcoI-BamHI-digested plasmid pET11d. The yjgR gene from E. coli was PCR-amplified with the following oligonucleotide primers: YJGR-FOR, 5′-ATC CCA TGG GTG AAC CCC TGT TAA TTG CCC GC; and YJGR-REV, 5′-GGA TCC CTT TAT TAT TGT TAG CAA AGT GTG CTT CG. Plasmid pYjgR was constructed in the same manner as plasmids pRpiB-RRN and pRpiB-RQR.
Plasmid pCH201, a derivative of plasmid pACYC184 containing an ssrA gene that encodes an ANDH6D peptide tag, was constructed by cassette mutagenesis with BsaI-digested plasmid pKW8 as described (16). Plasmid pCH403 is a derivative of pER203 (17), which contains an ssrA gene that encodes a ANDH6D peptide tag in addition to the argW gene (encoding tRNA5Arg) under the control of its own promoter. Plasmid pCH403 was constructed by first subcloning an NcoI-EcoRI fragment containing ssrA from pCH201 into pER203 to generate plasmid pCH402. A DNA fragment containing the argW gene and its promoter was then PCR-amplified by using the following primers: ARGW-FOR, 5′-AAA GCA GAA GCG CGC CAA AAA GCA GAA CGT GCG; and ARGW-REV, 5′-GAA GCG AAC CAT GAC GAA CTG TAA ATC TAC GG. This fragment was digested with BssHII, and ligated into plasmid pCH402 cut with BssHII and XmnI. The identities of all gene sequences and mutations were confirmed by DNA sequencing.
SsrA Tagging in Vivo.
Strains of E. coli X90 ssrA∷cat and X90 ssrA∷cat (DE3) containing plasmid pCH201 (or pCH403) and a protein expression plasmid were grown overnight at 37°C in LB medium supplemented with 100 μg/ml ampicillin and 20 μg/ml tetracycline. Fresh cultures were started the next day by diluting the overnight culture to an OD600 of 0.4 in fresh LB medium supplemented with 100 μg/ml ampicillin and 20 μg/ml tetracycline. After growth for 1.0 h at 37°C, the protein synthesis from Ptrc or T7 promoters was induced by adding IPTG to a final concentration of 1 mM followed by further culture for 2 h. Samples (1.0 ml) were removed and collected by centrifugation; the supernatant was removed and the cell pellet was immediately frozen at −80°C.
Frozen bacterial pellets for Western analysis were lysed in 50–100 μl of urea lysis buffer (8 M urea/100 mM NaCl/10 mM Tris⋅HCl, pH 8.0) with vigorous vortexing, and urea-soluble total protein was collected as the supernatant after centrifugation in a microcentrifuge at top speed for 10 min. Total protein concentration was estimated by Bradford total protein assay (Bio-Rad), and equal amounts of total urea-soluble protein (≈10–15 μg) from each strain were resolved by 10% Tris–N-tris(hydroxymethyl)glycine SDS/PAGE and the gels transferred to Immobilon-P (Millipore) polyvinylidene fluoride membranes by using standard Western-blotting procedures. SsrA(ANDH6D)-tagged proteins were detected by using His-probe H15 (Santa Cruz Biotechnology) with ECL+plus Western-blotting detection reagent (Amersham Pharmacia) and x-ray film.
Purification of RbsK-SsrA(ANDH6D) Fusion Peptides.
To identify the sites of SsrA tagging in ribokinase, 50-ml cultures of the appropriate X90 ssrA∷cat strains were started at an OD600 of 0.1 in LB medium supplemented with 100 μg/ml ampicillin and 20 μg/ml tetracycline from overnight cultures. At an OD600 of ≈1.0, the synthesis of ribokinase was induced by adding IPTG to a final concentration of 1 mM, followed by further incubation at 37°C with aeration for 2.5 h. The cultures were harvested, frozen at −80°C, and lysed in 12 ml of urea lysis buffer. SsrA(ANDH6D)-tagged ribokinase was purified by Ni2+-nitrilotriacetic acid (NTA) (Qiagen, Chatsworth, CA) affinity chromatography by using urea-lysis buffer as the wash buffer. SsrA(ANDH6D)-tagged ribokinase was eluted with 1.6 ml of elution buffer (2 M urea/100 mM acetic acid), the eluate neutralized with 80 μl of 2 M Tris⋅HCl (pH 9.5), and digested with 0.04 mg/ml of protease V8 (Worthington Biochemicals) overnight at 30°C. Under these conditions, cleavage was restricted to Glu-Xaa sequences. RbsK-SsrA(ANDH6D) fusion peptides were purified by a second round of Ni2+-NTA affinity chromatography, and eluted from the resin with 0.06% trifluoroacetic acid.
Identification and Quantitation of RbsK-SsrA(ANDH6D) Fusion Peptides.
Purified RbsK-SsrA(ANDH6D) fusion peptides were identified at the MIT Biopolymers Laboratory by liquid chromatography-electrospray ionization mass spectrometry by using a Perkin–Elmer Sciex model 365 triple-stage mass spectrometer attached to an Applied Biosystems model 140 C HPLC system. Peptide identification was confirmed by N-terminal sequence analysis of HPLC-purified fusion peptides. Peptides were purified/analyzed by reverse-phase HPLC on a 3.9 × 300 mm C18 column (Vydac, Hesperia, CA) by using the following gradient conditions: 0–5 min, 100% buffer A; 5–50 min, 0–35% buffer B; 50–80 min, 35–100% buffer B; where buffer A is 0.06% aqueous trifluoroacetic acid, and buffer B is 0.052% trifluoroacetic acid in 80% acetonitrile. Peptides were detected by UV absorption at 214 nm, and peptide quantitation was performed with SHIMADZU CLASS-VP software (v.5.03) to determine the area of HPLC chromatogram peaks corresponding to each RbsK-SsrA(ANDH6D) fusion peptide.
Results
Identification of SsrA Tagging Sites in Ribokinase.
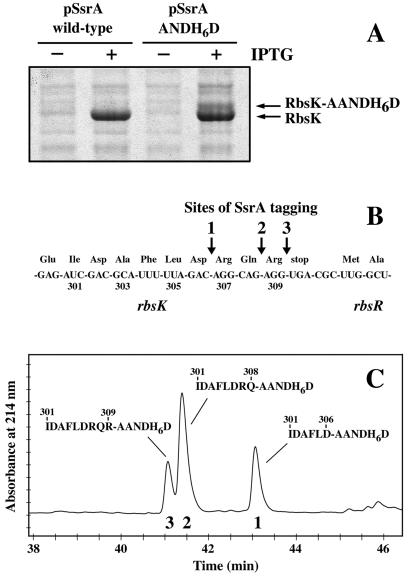
E. coli ribokinase is tagged by the SsrA system at a site that maps roughly to the last few residues of this protein (9). To identify this site more precisely, we allowed tagging to occur in an ssrA− strain bearing one plasmid (pRbsK1) containing the rbsK gene under Ptrc promoter control and a portion of the downstream rbsR gene and a second plasmid (pCH201) containing an SsrA mutant that encodes an ANDH6D peptide tag. This mutant tag prevents degradation, can be detected immunochemically, and allows affinity purification of SsrA-tagged proteins. In cells carrying both pRbsK1 and pCH201, approximately 10–25% of expressed ribokinase was tagged near its C terminus as assayed by Coomassie blue-stained SDS-polyacrylamide gels (Fig. 1A).
Figure 1.
Identification of SsrA(ANDH6D)-tagging sites in RbsK. (A) RbsK was expressed from the Ptrc promoter with (+) or without (−) IPTG induction in cells containing either wild-type SsrA or SsrA(ANDH6D). Cell lysates were electrophoresed on SDS-polyacrylamide gels and stained with Coomassie blue. The positions of full-length RbsK and SsrA(ANDH6D)-tagged RbsK are indicated. Densitometry indicated that as much as 25% of total RbsK may be tagged. (B) The nucleotide sequence encoding the 3′ end of wild-type rbsK and the 5′ end of rbsR with the corresponding amino acid sequence (in three letter code) is depicted. Positions of SsrA(ANDH6D) tagging in ribokinase are indicated by arrows. (C) Reverse-phase HPLC chromatogram of Ni2+-NTA-purified SsrA(ANDH6D)-RbsK fusion peptides. Electrospray mass spectrometry gave masses of 2001.87 Da for peptide 1, 2286.03 Da for peptide 2, and 2442.38 Da for peptide 3. The calculated mass for the sequences shown are 1 (2001.64 Da), 2 (2286.03 Da), and 3 (2442.12 Da). The N-terminal sequences of peptides 1 and 2 were I-D-A-F-L-D-A-A-N-D-H-H-H-H-H and I-D-A-F-L-D-R-Q-A-A-N-D-H-H-H, respectively.
SsrA(ANDH6D)-tagged ribokinase was purified by Ni2+-NTA affinity chromatography and digested with V8 protease; peptides were purified by an additional step of Ni2+-NTA chromatography. Coupled HPLC-electrospray mass spectrometry analysis showed three major species, each with a mass expected for a different junction peptide between RbsK and the AANDH6D tag, with tagging after RbsK residues Asp-306, Gln-308, and Arg-309 (Fig. 1 B and C). The identities of the former two peptides were confirmed by N-terminal sequencing (see legend to Fig. 1). These sites of SsrA tagging correspond to ribosome stalling at the codons for Arg-307 (AGG), Arg-309 (AGG), and the opal termination codon (UGA) (Fig. 1B). Assuming that each peptide was recovered with equal yield, roughly 24–29% of the tagging occurred at the position before Arg-307, 49–53% before Arg-309, and 18–27% before the opal stop codon (Fig. 1C and Fig. 2B). Taken together, these results show that translation of rbsK mRNA is completed normally only 75–90% of the time. In the remaining instances, the SsrA system intervenes, resulting in tagging at one of three closely spaced positions at or near the protein C terminus. RbsK overexpression is required for fine analysis of tagging, and the distribution of sites and/or overall tagging levels could change with the level of expression.
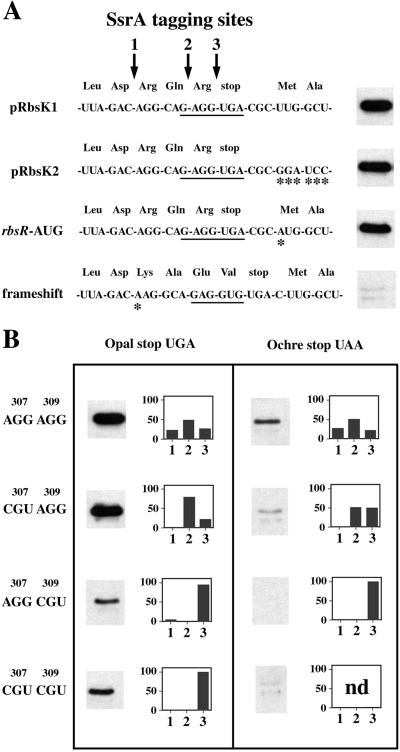
Figure 2.
Tagging of ribokinase expressed mutants of rbsK. (A) Nucleotide and protein sequences of the rbsK-rbsR region from four different ribokinase expression plasmids. The underlined nucleotides are complementary to the 3′ end of 16S rRNA and represent the presumed rbsR ribosome-binding site. Asterisks (*) indicate nucleotide changes relative to pRbsK1. All ribokinase expression plasmids except pRbsK2 (which contains no rbsR sequence) contain 115 bp of downstream rbsR sequence. To the right of each sequence is a Western blot whose intensity is proportional to the level of SsrA(ANDH6D) tagging of RbsK in a strain containing that expression plasmid. (B) Western blots of tagged RbsK and histograms of the relative distribution of tagging sites for each mutant construct. The identity of codons 307 and 309 are shown to the right, and the identity of the stop codon is shown above each panel. Tagging sites 1, 2, and 3 correspond to those shown in A. nd, not determined.
SsrA Tagging of rbsK Mutants.
Three mechanisms may contribute to SsrA tagging of ribokinase: (i) interference with completion of ribokinase translation by ribosomes initiating translation of the downstream rbsR mRNA (Fig. 1B); (ii) ribosome pausing at the rare AGG arginine codons encoding Arg-307 and Arg-309; and (iii) inefficient translation termination at the opal stop codon. To assess the importance of these potential mechanisms, we constructed mutations in rbsK and determined the corresponding level and sites of SsrA(ANDH6D) tagging (Fig. 2). One series of rbsK mutations included a frameshift mutation that places the AGG codons out of frame, a mutation that changes the rbsR start codon from UUG to AUG, and a construct (pRbsK2) that lacks the downstream rbsR sequence found in pRbsK1 (Fig. 2A). Another set of mutants contained synonymous arginine mutations in which the rare AGG codons were substituted singly or doubly with CGU, the most common Arg codon (Fig. 2B). Finally, the opal (UGA) termination codon of rbsK was changed to an ochre (UAA) stop codon singly and in combination with the AGG → CGU mutations (Fig. 2B). The level of ribokinase expressed from all constructs was equivalent as determined by SDS/PAGE analysis of urea-soluble cell extracts (data not shown).
No significant change in the overall level (Fig. 2A) or distribution (data not shown) of RbsK tagging was observed for the pRbsK2 and rbsR start codon → AUG constructs. These results suggest that the initiation of RbsR translation has little or no effect on RbsK tagging. One caveat is that both the pRbsK2 and the rbsR start codon → AUG constructs still contain the rbsR ribosome-binding site (Fig. 2A), which might interfere with ribokinase translation. To test this model, ribokinase was expressed from a frameshift construct that maintains the ribosome-binding site of rbsR but alters the 3′ rbsK reading frame (Fig. 2A). Ribokinase from this construct was tagged to a much lower extent than ribokinase from pRbsK1 (Fig. 2A). We conclude that ribosomes initiating translation of RbsR or binding to the rbsR Shine–Dalgarno sequence have little or no effect on SsrA(ANDH6D) tagging of ribokinase.
Replacing the AGG codons at the end of the rbsK gene with synonymous codons had different effects that depended on the mutated position(s). The Arg-307 AGG → CGU mutant showed little or no change in the overall level of tagging as determined by Western-blot analysis, although tagging was no longer detected at the site corresponding to Arg-307 (Fig. 2B). Ribokinase from the single Arg-309 AGG → CGU mutant and the double Arg-307/Arg-309 AGG → CGU mutant was tagged at approximately 50% the level of wild type (Fig. 2B). The Arg-309 → CGU mutation not only eliminated tagging at the Arg-309 site, but also dramatically reduced tagging at the upstream Arg-307 position (Fig. 2B). The double Arg-307/Arg-309 AGG → CGU mutant abolished all tagging except at the stop codon (Fig. 2B). These data support the possibility that ribosome pausing at both rare AGG arginine codons leads to SsrA tagging of ribokinase, but show that the AGG codon encoding Arg-309 is more effective in recruiting SsrA.
Opal stop codons (UGA) are generally less efficient as translational terminators than are ochre stop codons (UAA) (18). Ribokinase expressed from an rbsK variant containing an ochre termination codon (UAA) in place of the normal opal codon showed a 3- to 4-fold reduction in tagging, although the distribution of tagging sites was essentially unchanged (Fig. 2B). Tagging was further decreased by combining the ochre stop codon with the AGG → CGU mutations, with the Arg-309 → CGU mutation having a greater effect (Fig. 2B). The triple mutant containing the ochre stop and both AGG → CGU changes showed only very low levels of tagging (Fig. 2B). The nucleotide immediately after the stop codon seems to help determine stop-codon efficiency, and UGAC is thought to be a very inefficient stop signal (18, 19). Consistent with this idea, changing the rbsK terminator from UGAC to UGAU eliminated virtually all tagging in the context of the Arg-307/Arg-309 AGG → CGU mutations (data not shown). We conclude that the efficiency of the rbsK translation termination codon affects ribokinase tagging, with less efficient stop signals causing higher levels of tagging.
Multiple Copies of argW (tRNA5Arg) Suppress RbsK Tagging.
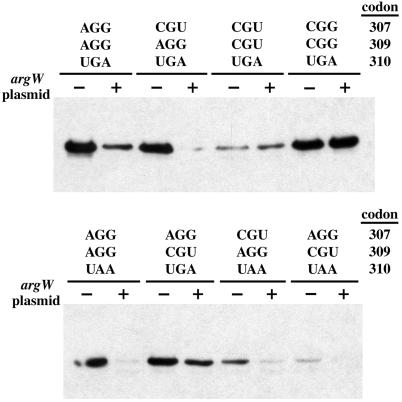
AGG codons are decoded by tRNA5Arg, which is encoded by argW and is one of the least abundant tRNA species in E. coli (20). When the argW gene was placed on the same multicopy plasmid as ssrA(ANDH6D), the overall level of RbsK tagging was reduced, usually significantly, for all genes containing AGG codons (Fig. 3). By contrast, tRNA5Arg suppression of SsrA(ANDH6D) tagging was not observed for constructs in which Arg-307 and Arg-309 were encoded by CGU or by CGG codons (Fig. 3). Analysis of RbsK-SsrA(ANDH6D) junction peptides from strains containing the suppression plasmid showed tagging at stop codons and at CGG codons, which are also considered rare (20), but not at any AGG codons (data not shown). We conclude that limiting tRNA5Arg generally leads to ribosome stalling and subsequent SsrA tagging at AGG codons.
Figure 3.
Overexpression of argW suppresses SsrA(ANDH6D) tagging at AGG codons. Anti-His6 Western-blot analysis of SsrA(ANDH6D)-tagged RbsK from a wild-type strain (−) or a strain (+) that overexpresses tRNA5Arg from a plasmid containing the argW gene. The identity of rbsK codons 307, 309, and 310 are indicated.
Rare Arg-Codon/Stop-Codon Pairs Are General Tagging Determinants.
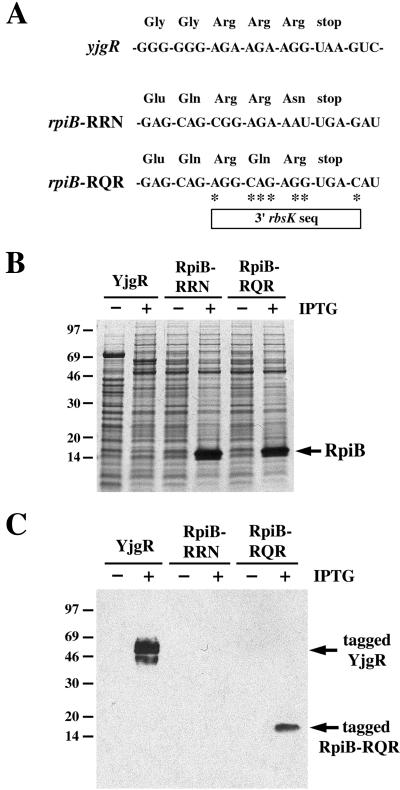
Because efficient SsrA tagging of ribokinase seems to be caused by rare AGG arginine codons in proximity to the stop codon, we sought to determine whether other E. coli genes with rare arginine codons near the stop codon give rise to SsrA-tagged proteins. The yjgR gene ends with three consecutive rare arginine codons (AGA AGA AGG UAA) (Fig. 4A). Western-blot analysis showed that overexpressed YjgR protein was tagged by SsrA(ANDH6D) at a position near its C terminus (Fig. 4C). We next tested the rpiB gene, which has two consecutive rare arginine codons separated from the stop codon by an asparagine codon (CGG AGA AAU UGA) (Fig. 4A). In contrast to RbsK and YjgR, overexpressed RpiB was tagged by SsrA(ANDH6D) at a level too low to detect in the experiment shown in Fig. 4C. We next constructed an rpiB variant in which the last three codons and the stop signal were replaced by the corresponding rbsK sequence (Fig. 4A). This RpiB-RQR variant was tagged at a much higher level than protein from the wild-type rpiB gene (Fig. 4C), even though both proteins were expressed at comparable levels (Fig. 4B). This result shows that the 3′ rbsK sequence is sufficient to direct efficient SsrA tagging in other genetic contexts. Overall, these results suggest that SsrA tagging at positions corresponding to rare arginine codons is much more efficient when these codons are immediately adjacent to the stop codon, as in rbsK and yjgR.
Figure 4.
The 3′ end of the rbsK gene directs SsrA tagging in a different context. (A) The nucleotide sequences and corresponding amino acid sequence for the 3′ end of yjgR, the wild-type rpiB gene, and an rpiB mutant containing the 3′ end of the rbsK gene. Asterisks (*) indicate the position of mutated nucleotides relative to wild-type rpiB. (B) Total urea-soluble protein from IPTG-induced and uninduced cells carrying these constructs was resolved by SDS/PAGE, followed by staining with Coomassie blue. The migration position of RpiB is indicated. YjgR has a predicted Mr of 54 kDa. (C) Anti-His6 Western-blot analysis of SsrA(ANDH6D)-tagged YjgR and SsrA(ANDH6D)-tagged RpiB.
Discussion
E. coli ribokinase is modified by the SsrA system at three major positions (Fig. 1B). These sites correspond to tag addition after Asp-306, Gln-308, and the C-terminal residue, Arg-309. If SsrA tagging occurs when ribosomes pause at codons that are translated slowly, then pausing is predicted at the rare AGG codon encoding Arg-307, the rare AGG codon encoding Arg-309, and the UGA stop codon. Studies from this laboratory have revealed SsrA tagging at positions that would correspond to stalling at artificial clusters of rare Arg codons and at the stop codons of the mRNAs encoding λ repressor (UGA) and YbeL (UAA) (9, 17). Hence, a growing body of evidence suggests that translational pausing at certain rare codons and some stop codons provides an opportunity for SsrA tagging of many different bacterial gene products.
Mutation of the rare AGG codons and/or the stop codon in the rbsK mRNA affected SsrA tagging in several ways that support the idea that ribosome pausing at these sites is required for the tagging reaction. First, replacing either or both rare codons with common Arg codons eliminated tagging at the corresponding site(s). Overexpression of tRNA5Arg (argW), which decodes AGG, also eliminated tagging at these sites. Second, when both Arg residues were encoded by common codons, tagging of the intact protein was still observed when the wild-type UGAC stop signal was present, but no significant tagging was observed for rbsK mRNAs with the more efficient UAAC or UGAU stop signals (18, 19). In each of these cases, mutations or conditions that increase the efficiency of ribosomal transactions reduce tagging, as expected for the pausing model.
In several instances, mutations at one codon in rbsK had effects on tagging at positions corresponding to upstream codons. For example, replacing the UGA termination codon with UAA reduced tagging before the stop codon but also reduced tagging before both rare AGG codons (Fig. 2B). Similarly, with the wild-type opal terminator or the mutant ochre stop, mutating the Arg-309 codon from AGG to CGU eliminated tagging before this site but also dramatically reduced tagging before Arg-307 (Fig. 2B). These results suggest that molecular events influenced by the stop codon and the terminal Arg codon are able to affect ribosome pausing at upstream codons. One model explains these upstream effects: tRNA5Arg becomes sequestered at the Arg-309 codon in the P site of the ribosome during the extended pause required for release factor-2 to decode the UGAC stop signal. This sequestration depletes the pool of tRNA5Arg, which is already small, thereby increasing the frequency of pausing and hence SsrA tagging before AGG codons (307 and 309) on rbsK mRNAs bound to other ribosomes. This sequestration model correctly predicts that increasing the efficiency of translation termination reduces tagging before both AGG sites in rbsK. This model also predicts that a common Arg-309 codon will cause decreased tagging before Arg-307, even if this latter residue is encoded by a rare AGG codon. A reciprocal effect of 307 on 309 is not expected if translation of the 308 codon is fast because tRNA5Arg would not be sequestered efficiently.
Our results indicate that completion of RbsK translation is sufficiently slow under the conditions assayed to cause 10–25% of the newly translated protein chains to be tagged by SsrA(ANDH6D). Tagging of RbsK by wild-type SsrA should also occur at a similarly high level and would result in degradation of the tagged protein chains. At first glance, this process of inefficient completion of translation followed by tagging and degradation seems to be a profligate use of cellular resources. Indeed, the last five residues of RbsK are not required for activity in vivo (unpublished results). Why not truncate the protein or synthesize the same RbsK sequence more economically by using common Arg codons for positions 307 and 309 and by using an efficient termination codon? One answer may be that the wild-type DNA sequence at the 3′ end of rbsK plays some additional function. For example, ribosomal pausing at this site might regulate the closely spaced rbsR gene, although we were unable to detect an effect of SsrA on RbsR expression (unpublished results). Another intriguing possibility is that the wild-type rbsK sequence has evolved to allow down-regulation of RbsK expression after translation has begun. In this scenario, we envision that the intracellular levels of tRNA5Arg and/or release factor-2 might themselves be regulated downward under some growth conditions, leading to a large reduction in the efficiency of completion of RbsK translation with concomitant SsrA tagging and degradation. In fact, tRNA5Arg levels are subject to growth-rate regulation via the FIS transcription factor (21, 22) and release factor-2 activity is also subject to regulation at the levels of expression and modification (23, 24).
The combination of a rare arginine codon and an adjacent inefficient terminator seems to be a general determinant of SsrA tagging. In this regard, we note that 24 protein genes in E. coli end with AGG and 28 end with AGA, another rare arginine codon. The overall frequencies of usage for these rare codons in E. coli are 0.12% and 0.21%, respectively. On the basis of these frequencies, only about 16 genes would be expected to end with AGG or AGA, whereas 52 are observed. This overrepresentation of rare codons at the 3′ ends of E. coli genes has been noted and attributed, in part, to the requirements of closely spaced downstream genes for translation initiation signals (25). On the basis of the discussion in the preceding paragraph, another possibility is that the proteins encoded by these genes are subject to some type of global regulation mediated by way of translation termination and the SsrA system.
Acknowledgments
We thank Graham Walker, Peter Chivers, Sean Moore, and Tom RajBhandary for helpful discussions. This work was supported by Grant AI-16892 from the National Institutes of Health. C.S.H. was supported by a fellowship (DRG 1686) from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation.
Abbreviations
- RbsK
E. coli enzyme ribokinase
- RpiB
ribose-5-phosphate isomerase
- IPTG
isopropyl β-d-thiogalactoside
- NTA
Ni2+-nitrilotriacetic acid
References
- 1.Keiler K C, Waller P R, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 2.Karzai A W, Roche E D, Sauer R T. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 3.Muto A, Ushida C, Himeno H. Trends Biochem Sci. 1998;23:25–29. doi: 10.1016/s0968-0004(97)01159-6. [DOI] [PubMed] [Google Scholar]
- 4.Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A. J Mol Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- 5.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. Proc Natl Acad Sci USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu G F, Reid G E, Zhang J G, Moritz R L, Simpson R J. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 7.Herman C, Thevenet D, Bouloc P, Walker G C, D'Ari R. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roche E D, Sauer R T. J Biol Chem. 2001;76:28509–28515. doi: 10.1074/jbc.M103864200. [DOI] [PubMed] [Google Scholar]
- 10.Abo T, Inada T, Ogawa K, Aiba H. EMBO J. 2000;19:3762–3769. doi: 10.1093/emboj/19.14.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranquet C, Geiselmann J, Toussaint A. Proc Natl Acad Sci USA. 2001;98:10220–10225. doi: 10.1073/pnas.171620598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Retallack D M, Johnson L L, Friedman D I. J Bacteriol. 1994;176:2082–2089. doi: 10.1128/jb.176.7.2082-2089.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Retallack D M, Friedman D I. Cell. 1995;83:227–235. doi: 10.1016/0092-8674(95)90164-7. [DOI] [PubMed] [Google Scholar]
- 14.Iida A, Harayama S, Iino T, Hazelbauer G L. J Bacteriol. 1984;158:674–682. doi: 10.1128/jb.158.2.674-682.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hope J N, Bell A W, Hermodson M A, Groarke J M. J Biol Chem. 1986;261:7663–7668. [PubMed] [Google Scholar]
- 16.Williams K P, Martindale K A, Bartel D P. EMBO J. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche E D, Sauer R T. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tate W P, Mannering S A. Mol Microbiol. 1996;21:213–219. doi: 10.1046/j.1365-2958.1996.6391352.x. [DOI] [PubMed] [Google Scholar]
- 19.Pavlov M Y, Freistroffer D V, Dincbas V, MacDougall J, Buckingham R H, Ehrenberg M. J Mol Biol. 1998;284:579–590. doi: 10.1006/jmbi.1998.2220. [DOI] [PubMed] [Google Scholar]
- 20.Dong H, Nilsson L, Kurland C G. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson L, Emilsson V. J Biol Chem. 1994;269:9460–9465. [PubMed] [Google Scholar]
- 22.Emilsson V, Nilsson L. J Biol Chem. 1995;270:16610–16614. doi: 10.1074/jbc.270.28.16610. [DOI] [PubMed] [Google Scholar]
- 23.Adamski F M, McCaughan K K, Jorgensen F, Kurland C G, Tate W P. J Mol Biol. 1994;238:302–308. doi: 10.1006/jmbi.1994.1293. [DOI] [PubMed] [Google Scholar]
- 24.Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyre-Walker A. J Mol Evol. 1996;42:73–78. doi: 10.1007/BF02198830. [DOI] [PubMed] [Google Scholar]