Abstract
The insect adipokinetic hormones (AKHs) are a large family of peptide hormones that are involved in the mobilization of sugar and lipids from the insect fat body during energy-requiring activities such as flight and locomotion, but that also contribute to hemolymph sugar homeostasis. Here, we have identified the first insect AKH receptors, namely those from the fruitfly Drosophila melanogaster and the silkworm Bombyx mori. These results represent a breakthrough for insect molecular endocrinology, because it will lead to the cloning of all AKH receptors from all model insects used in AKH research, and, therefore, to a better understanding of AKH heterogeneity and actions. Interestingly, the insect AKH receptors are structurally and evolutionarily related to the gonadotropin-releasing hormone receptors from vertebrates.
Insects constitute the largest animal group on earth and are economically and ecologically extremely important, because most flowering plants depend on insects for their pollination, and insects can be serious pests. Despite the importance of insects, however, our knowledge of their endocrinology is still incomplete. Although in the last 20 years considerable progress has been made with the isolation and identification of peptide hormones from insects (1, 2), the identification of their receptors has remarkably lagged behind (1–4). The adipokinetic hormones (AKHs) are one of the best studied insect neurohormones with more than 30 different family members isolated from over 70 species (1, 2, 4–11). The action of AKH is comparable to that of glucagon from mammals. It contributes to hemolymph sugar homeostasis, but it is also involved in the mobilization of sugar and lipids from the fat body during energy-requiring activities, such as flight or locomotion (1, 2, 4–11). Here we describe the identification of an AKH receptor from the fruitfly Drosophila melanogaster and that from another model insect, the silkworm Bombyx mori. These findings will provide an important lead to find additional insect AKH receptors, which will help us to understand AKH heterogeneity and actions.
Materials and Methods
Extraction of the Receptor Ligand.
A total of 400 g of third-instar larvae from D. melanogaster (Canton S.) were ground to powder under liquid nitrogen, boiled in 3 vol of deionized water for 20 min, and cooled to 0°C. After acetic acid addition (final pH, 3.0) and homogenization with a Braun food processor, the mixture was centrifuged, and the supernatant was brought to pH 7.0 with a diluted NaOH solution. The extract was then desalted by using several SepPak C18 cartridges (Waters). After being rinsed with 5 ml of H2O, each cartridge was eluted with 4 ml of 50% acetonitril in 0.1% trifluoroacetic acid. All eluates were lyophilized and used as a starting material for HPLC (Table 1).
Table 1.
HPLC purification of the Drosophila GnRHR receptor ligand
| Column | Gradient of acetonitrile in 0.1% trifluoroacetic acid | Retention time of the peptide, min |
|---|---|---|
| 1. Spherisorb C-18 (20 × 250 mm; 5 μm; 80 Å) | 10–60% (40 min; 8 ml/min) | 27.0–28.0 |
| 2. Partisil ODS-3 (4 × 250 mm; 5 μm; 60 Å) | 10–60% (30 min; 1 ml/min) | 17.0–18.0 |
| 3. LiChrospher RPSelect (4 × 250 mm; 5 μm; 60 Å) | 20–50% (30 min; 1 ml/min) | 16.5–16.8 |
| 4. Nucleosil C-18 (4 × 250 mm; 5 μm, 300 Å) | 20–30% (40 min; 1 ml/min) | 19.06 |
| 5. Spherisorb C-18-2 (4 × 250 mm; 5 μm; 80 Å) | 20–40% (40 min; 1 ml/min) | 20.66 |
| 6. Nucleosil C-18 (4 × 250 mm; 5 μm; 120 Å) | 20–30% (40 min; 1 ml/min) | 25.55 |
| 7. Spherisorb C-18-2 (4 × 250 mm; 5 μm; 80 Å) | 20–30% (40 min; 1 ml/min) | 20.91 |
The starting material was an aqueous extract from 400 g of third-instar larvae (see Materials and Methods).
HPLC of the Extracts.
The HPLC system used was from Shimazu (LC-6A; SPD-6AV; SCL-6B; C-R6A). Columns 1, 2, 5, and 7 (see Table 1) were purchased from Latek (Heidelberg), columns 4 and 6 were purchased from Macherey–Nagel (Düren, Germany), and column 3 was purchased from Merck.
Cell Culture, Transfection, and Creation of Cell Line CHO/G16/PCG.6.
Chinese hamster ovary (CHO) cells, stably expressing the α subunit of the human G protein G16 (CHO/G16) (12) were a kind gift from S. Rees and J. Stables (Glaxo Wellcome, Stevenage, U.K.), and were maintained in CHO medium (DMEM-F12 supplemented with 5% FBS; all reagents from Life Technologies) and selected with Hygromycin B (200 μg/ml; Life Technologies). Primers were designed based on sequence information from the Drosophila gonadotropin-releasing-hormone (GnRH) receptor (13), recognizing the sequence surrounding the ATG start codon (5′-GGTACCAAAATGGCAAAAGTAGCTGAG-3′) (the last part of this sequence is corresponding to nucleotide positions 1–18 of figure 2 from ref. 13) and the stop codon (5′-GAATTCTTACTTCTGGCGGATCGG-3′) (the last part of this sequence is corresponding to nucleotide positions 1314–1331 of figure 2 from ref. 13) and used to amplify a full-length cDNA from third-instar larvae of D. melanogaster. A PCR product of 1,347 bp was cloned into a pcDNA3 expression vector (Invitrogen), and several independent clones were sequenced to verify the correct amplification of the cDNA. For stable transfection of the CHO/G16 cells with the vector construct, FuGene6 from Boehringer Mannheim was used as a transfection agent (according to the manufacturer's instructions). After antibiotic (1 mg/ml G418) and clonal selection, one cell clone (CHO/G16/PCG.6) of 10 original clones, which showed the highest response in our bioluminescence assay (see below), was chosen for further studies. Transient transfection of CHO/G16/PCG.6 with mitochondrially targeted aequorin/pcDNAI (Molecular Probes) was performed as follows: CHO/G16/PCG.6 cells were plated out in 6-well plates at about 60% confluency. After 24 h they were transfected with 0.8 μg of DNA per well by using 2.5 μl of FuGene6. Twenty-four hours after transient transfection, the cells were transferred to 96-well plates at a confluency of about 80%.
Bioluminescence Assay.
Bioluminescence assays were performed 48 h after the transient transfection in white 96-well microtiter plates using cells that were about 80–95% confluent. To reconstitute the holoenzyme apoaequorin, cells were incubated in CHO medium containing 0.1% FBS and 5 μM coelenterazine (Molecular Probes) for 3 h (12). Test compounds were dissolved in PBS and warmed up to 37°C, and 100 μl was added to the wells (final volume, 200 μl). Luminescence was measured at 37°C, by using a Wallac Victor2. The synthetic peptides were >95% pure and either synthesized by Genemed Synthesis [San Francisco; D. melanogaster AKH (Drm–AKH); Manduca sexta AKH (Mas–AKH); drostatin-A1), or purchased from Bachem (Bubendorf, Switzerland; Heliothis zea hypertrehalosaemic hormone (Hez–HrTH); Schistocerca gregaria AKH-II (Scg-AKH-II); corazonin].
Mass Spectrometry.
The positive ion electrospray ionization (ESI) mass spectra were recorded on a quadrupole-time-of-flight mass spectrometer (Micromass, Manchester, U.K.) equipped with a nano-ESI ion source. In the collision-induced dissociation (CID) experiments, the precursor ions were mass selected by the quadrupole and then accelerated into the hexapole collision cell (collision energy, 25 eV). Argon was used as a collision gas at an indicated manifold pressure of 8.7 × 10−5 mbar (1 bar = 100 kPa). The product ions formed by the internal excitation and subsequent dissociation of the parent ions were then mass analyzed by the orthogonal TOF analyzer.
Cloning of the Bombyx AKH Receptor.
Drosophila AKH receptor (AF077299) and Drosophila G protein-coupled receptor CG10698 (www.flybase.org) were aligned, and degenerate primers were designed from conserved regions. The primers used were as follows: sense, 5′-GGG/TG/CITA/GIACIGTICAA/GTGGC/T-3′ (corresponding to nucleotide positions 291–318 of figure 2 from ref. 13); and antisense, 5′-A/GTAA/GTAIGGIGTCCAA/GCAIATIAIA/G-3′ (corresponding to nucleotide positions 811–840 of figure 2 from ref. 13), where I is deoxyinosine; primers were used on cDNA from fifth-instar whole B. mori larvae. The PCR parameters were as follows: 94°C for 3 min, then touchdown PCR for 7 cycles, 95°C for 30 s, 59°C for 45 s, decreasing 1°C for each cycle and 68°C for 90 s followed by 35 cycles of 95°C for 30 s, 52°C for 45 s, 68°C for 90 s, and a final extension step of 68°C for 10 min. pCR4-TOPO (Invitrogen) was used for the cloning and the SMART RACE cDNA kit (CLONTECH) was used for the rapid amplification of cDNA ends (RACE) reactions. The 3′-RACE reactions were made with the sense primer, 5′-AGAGCACCCCGAAGTTAAAGG-3′ (corresponding to nucleotide positions 768–788 of our GenBank database submission; accession no. AF403542) followed by the nested sense primer 5′-TCTTACGGCTCCCTGCCAACG-3′ (corresponding to nucleotide positions 805–825 of AF403542). The 5′-RACE reactions were made with the antisense primer, 5′-AAGACCAGGACTATGGTGACC-3′ (corresponding to nucleotide positions 1017–1037 of AF403542) followed by the nested antisense primer 5′-CAGAAGACCGATGCCGCTTCG-3′ (corresponding to nucleotide positions 964–984 of AF403542). The PCR program was 94°C for 3 min, then touchdown PCR for 12 cycles, 94°C for 30 s, 70°C for 30 s, decreasing 2°C every 2 cycles and 72°C for 3 min followed by 23 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 3 min, and a final extension step of 72°C for 7 min. pCR4-TOPO (Invitrogen) was used for the cloning.
Transfection of CHO/G16 Cells with the Bombyx Receptor DNA.
Primers were designed, recognizing the sequences surrounding the ATG start codon (5′-CGGGGTACCAAGATGGATATAGACGAGAAAGTG-3′; corresponding to nucleotide position 223–243 of AF403542) and the stop codon (5′-CCGCTCGAGTTAAACGATACCGTTCGTTACG-3′; corresponding to nucleotide position 1419–1440 of AF403542) of the Bombyx receptor cDNA and used to amplify a full-length cDNA from fifth-instar whole B. mori larvae. To transfect CHO/G16 cells with the DNA coding for the Bombyx receptor, a similar transfection procedure was carried out as with the Drosophila receptor, the only difference being that no clonal selection of the cells was performed.
Results and Discussion
We have previously cloned a Drosophila G protein-coupled receptor that was structurally and evolutionarily related to the three known mammalian glycoprotein hormone (gonadotropin and thyroxin stimulating-hormone) receptors (14). To find additional possible Drosophila glycoprotein hormone receptors, we screened, using the blast algorithm, the Drosophila Genome Project database (that at that time was 10–20% complete) with each of the seven transmembrane helices of the first Drosophila glycoprotein hormone receptor, which resulted in the cloning of a Drosophila G protein-coupled receptor that was structurally related to the vertebrate gonadotropin-releasing-hormone (GnRH) receptors (36% amino acid residue identity with the catfish and 31% with the rat GnRH receptor) (13). One intron in the Drosophila receptor gene occurred at the same position and had the same intron phasing as one intron in the rat GnRH receptor gene, showing that the two receptors were not only structurally related, but also evolutionarily related (13).
The Drosophila GnRH receptor-related (GnRHR) receptor is an orphan receptor, and its ligand is unknown, although we expected it to be related to one of the vertebrate GnRH peptides. To find the cognate Drosophila GnRHR receptor ligand, we stably expressed the receptor in CHO cells that were also stably expressing the α subunit of the “promiscuous” human G protein, G16 (12), and cloned one cell line (CHO/G16/PCG.6), expressing the receptor most abundantly. Two days before the assay, we transiently transfected these cells with a vector containing DNA coding for aequorin, and 3 h before the assay we added coelenterazine to the cell culture medium. Activation of the Drosophila GnRHR receptor in these pretreated cells would result in a Ca2+-induced bioluminescence response, which could easily be measured and quantified (12).
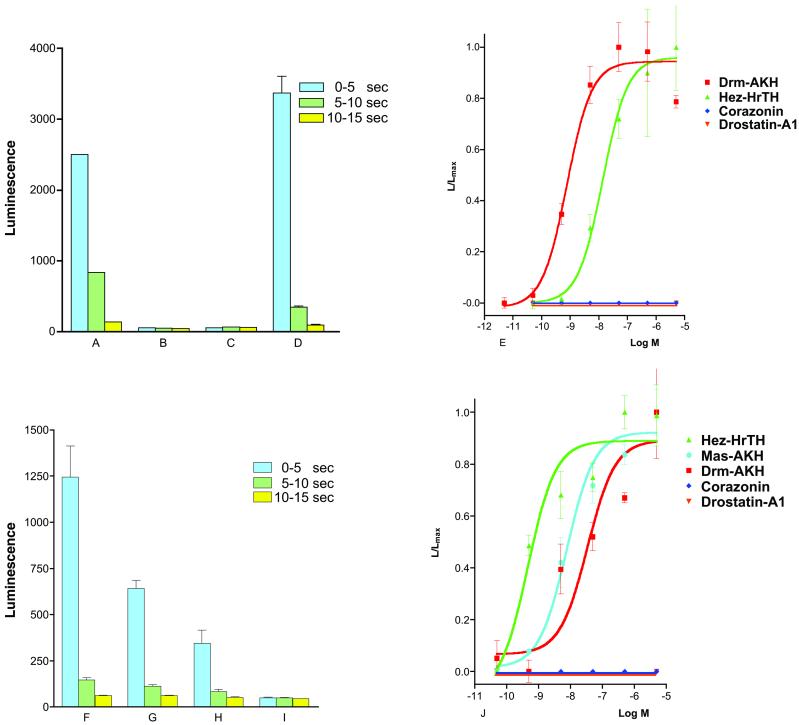
The Drosophila GnHR receptor is mostly expressed in third-instar larvae (13). We made, therefore, an aqueous extract from 400 g of third-instar larvae (about 4 × 105 animals) and investigated whether the extract contained the GnRHR receptor ligand, by using the bioluminescence response of the above-mentioned transformed CHO cells as a bioassay. This, indeed, turned out to be the case, which enabled us to purify the ligand by HPLC (Table 1). Fig. 1A gives an example of the bioluminescence response induced by the partially purified natural Drosophila GnRHR receptor ligand. Fig. 1B shows that CHO/G16 cells transfected with the empty vector did not react with the natural ligand. Furthermore, cells expressing the Drosophila GnRHR receptor did not react on addition of PBS alone (Fig. 1C). After seven HPLC purification steps (Table 1), the natural ligand was purified to apparent homogeneity, i.e., a single peak of the expected form.
Figure 1.
Bioluminescence responses of the cloned cell line CHO/G16/PCG.6 (PCG.6), expressing the previously published orphan receptor, Drosophila GnRHR receptor (13), of CHO/G16 cells, expressing the newly cloned Bombyx AKH receptor; and of a CHO/G16 cell line transfected with the empty vector. The vertical bars represent SEM, which are sometimes lower than the symbols used. In these cases, only the symbols are given. (A) Bioluminescence response of PCG.6 after addition of 1% of the active fraction in purification step 3 (Table 1). The responses 0–5, 5–10, and 10–15 s after addition of the fraction (total measured bioluminescence in these periods) are shown. Note that the bioluminescence response declines quickly, which is probably caused by receptor desensitization. (B) Response of CHO/G16 that was transfected with the empty vector, after addition of 1% of the active fraction. (C) Response of PCG.6 after addition of 100 μl of PBS. (D) Response of PCG.6 after addition of 10−6 M synthetic Drm–AKH. (E) Dose-response curves of the effects of synthetic Drm–AKH (red), Hez–HrTH (green), and the Drosophila neuropeptides corazonin (dark blue), and drostatin-A1 (orange) on PCG.6. Drm–AKH is the most potent peptide in causing a bioluminescence response (EC50, 8 × 10−10 M). (F) Response of CHO/G16 cells stably transfected with the Bombyx receptor (Fig. 3) after addition of 10−6 M Hez–HrTH. (G) Response of the same cells to 10−6 M Drm–AKH. (H) Response of these cells to 10−6 M of the locust peptide Schistocerca gregaria AKH-II. (I) Response of these cells to 100 μl PBS. (J) Dose-response curves of the effects of synthetic Hez–HrTH (green), Mas–AKH (light blue), Drm–AKH (red), corazonin (dark blue), and drostatin-A1 (orange) on the CHO/G16 cells transfected with the Bombyx receptor DNA.
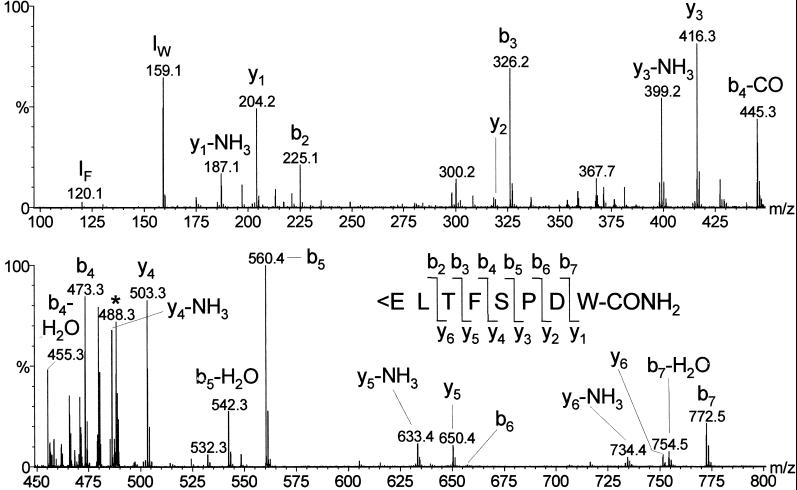
The structure of the purified ligand was determined by CID experiments using an electrospray mass spectrometer (Fig. 2). There was an abundant y3/b6 ion pair, showing enhanced cleavage of the peptide bond at the N-terminal side of proline (the “proline effect”) (15), but also other complementary ion pairs were observed: y6/b2, y5/b3, y4/b4, and y1/b7. The y2/b6 ions were much less abundant, because they resulted from cleavage of the peptide bond C-terminal from proline (15). The two N-terminal residues of the ligand were assigned after Drosophila database searches (www.flybase.org).
Figure 2.
CID spectrum of 0.3% of the material purified in step 7 of Table 1. The insertion gives the fragmentation pattern, showing that the purified peptide is identical to Drosophila adipokinetic hormone (Drm–AKH). <E means pyroglutamic acid. IF and IW are the immonium ions of Phe and Trp. The precursor ion [M + 2H]2+ at m/z 488.3 is marked with an asterisks. The mass of the neutral intact peptide is 974.6 Da. The CID spectrum of synthetic Drm–AKH was identical to that of the natural peptide.
The CID spectrum of Fig. 2 showed that the structure of the purified ligand was identical to that of a previously isolated, identified, and cloned Drosophila peptide, Drm–AKH (7, 16). Because our mass spectra suggested this structure, we synthesized Drm–AKH and compared the CID spectra from the natural ligand and synthetic Drm–AKH. This comparison showed that the two spectra were identical, confirming the proposed sequence of the Drosophila GnRHR receptor ligand.
Synthetic Drm–AKH was also tested on the transformed (CHO/G16/PCG.6) cells (Fig. 1D), showing that Drm–AKH gives a clear bioluminescence response indistinguishable from that of the natural ligand. Dose-response curves (Fig. 1E) showed that the bioluminescence responses induced by synthetic Drm–AKH have an EC50 of 8 × 10−10 M. Synthetic AKHs from other insect species (1, 4) also induced a bioluminescence response in the transformed cells, but with much less potency (e.g., hypertrehalosaemic hormone from the moth H. zea, Hez–HrTH; EC50, 2 × 10−8 M). Other neuropeptides, e.g., the Drosophila A-type allatostatins (17), did not activate the receptor. Even Drosophila corazonin, which has some structural features in common with the insect AKHs (18), did not give a response in the transformed CHO cells (Fig. 1E).
The above data, thus, clearly show that the cognate ligand of the Drosophila GnRHR receptor is Drm–AKH. These findings illustrate that it is dangerous to put names on orphan receptors based on structural and evolutionary relationships alone (“annotations”—they might, of course, be very useful in other contexts). Furthermore, our data represent a breakthrough for decades of work by other insect scientists to find or characterize insect adipokinetic hormone receptors (1, 3, 4, 19–22). Our results will now make it possible to clone all AKH receptors from all insects, and, because insect AKHs are structurally closely related to the red-pigment-concentrating hormone from crustaceans (AKH injected into crustaceans induce pigment concentration in chromatophores—red-pigment-concentrating hormone injected into insects induce lipid mobilization) (5, 23, 24), it will now also be possible to clone the crustacean red-pigment-concentrating hormone receptors.
From some insects it is known that they produce two or more different types of AKH (1, 2, 4, 6, 10), and it can be expected that these species have two or more different AKH receptors. In the present paper, we have only identified one Drosophila AKH receptor, but the Drosophila Genome Project database contains the sequence of a second G protein-coupled receptor (CG10698; www.flybase.org) that is closely related to the first Drosophila AKH receptor (now called Drm–AKH receptor-1) both with respect to amino acid sequence and gene structure (3). This receptor, therefore, is most likely to be a second Drm–AKH receptor, suggesting that many or perhaps all insect species have two or more AKH receptors.
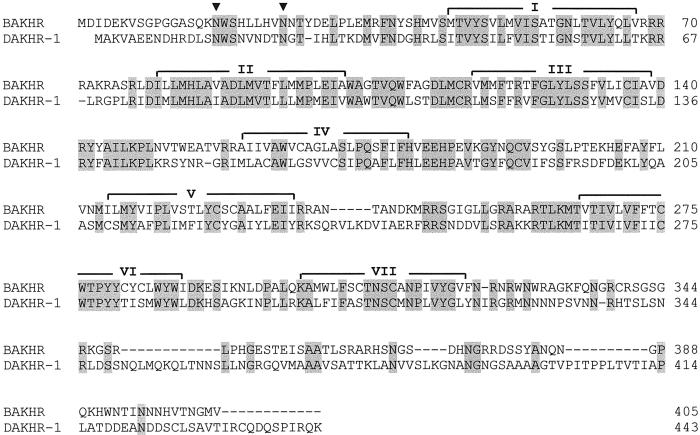
To illustrate the opportunities that our present findings offer, we have cloned an AKH receptor from another model insect, the silkworm B. mori (which belongs to a different insect order, the Lepidoptera, or moths and butterflies). This cloning was done by aligning the sequence of the Drm–AKH receptor-1 with that of the probable Drm–AKH receptor-2 (CG10698) and by using primers against their conserved regions, in conjunction with PCR and 3′/5′-RACE. Fig. 3 shows the primary structure of the cloned Bombyx receptor, which has 48% identical amino acid residues (68% conserved residues) in common with the Drm–AKH receptor-1. Furthermore, two potential glycosylation sites occur at the same positions within the two receptors (Fig. 3).
Figure 3.
Alignment of the B. mori AKH receptor (BAKHR) with the D. melanogaster AKH receptor (DAKHR-1, formally called Drosophila GnRHR receptor) (13). Identical amino acid residues are highlighted in gray, the potential N-glycosylation sites are indicated by filled triangles, and the seven membrane-spanning domains by I–VII. Dashed lines are spaces to optimize alignment. The complete nucleotide sequence of BAKHR has been deposited in the GenBank database with accession no. AF403542, and that of DAKHR-1 with accession no. AF077299.
We expressed the B. mori AKH (Bom-AKH) receptor in CHO/G16 cells and found that it is activated by low concentrations of a moth AKH peptide, the H. zea hypertrehalosaemic hormone (Hez–HrTH; EC50, 3 × 10−10 M) (Fig. 1 F and J). Hez–HrTH (25) has not been isolated from Bombyx so far, but another AKH peptide has been purified from this silkworm, which turned out to be identical to Mas–AKH, an AKH peptide originally isolated from the moth M. sexta (26). Mas–AKH also activated the Bom–AKH receptor, but with a lower affinity than Hez–HrTH (EC50, 8 × 10−9 M) (Fig. 1J). These results suggest that Bombyx has a second intrinsic AKH that is more related to Hez–HrTH than to Mas–AKH and that the Bom–AKH receptor is the high-affinity receptor for this second Bombyx AKH peptide. Drm–AKH did also activate the Bombyx receptor, but with a much lower potency than Hez–HrTH (EC50, 2 × 10−8 M), whereas other insect AKHs, such as the locust peptide Schistocerca–AKH-II, were less effective (Fig. 1 F–H and J). Corazonin did not stimulate the receptor, nor did other insect peptides that were unrelated to AKH, or PBS alone (Fig. 1 I and J). All of these data show that the Bombyx receptor is an AKH receptor that reacts to Bombyx and other moth AKHs with high affinity.
We have mentioned that the insect AKH receptors are structurally and evolutionarily related to the GnRH receptors from mammals. It is our own experience that evolutionarily related G protein-coupled receptors in different animal groups might have exchanged their ligands, but that their basic functional properties have roughly remained unchanged. The allatostatin receptors from insects, for example, are structurally clearly related to the somatostatin, galanin, and opioid receptors from mammals (27–32). Both the insect and the mammalian receptors are generally inhibitory receptors, a function that, thus, has been conserved, but their ligands are different in structure. Another example is that of the oxytocin/vasopressin receptor family, where the ligands have remained relatively similar during evolution (five of nine residues and a disulfide ring structure have been conserved) (33). These receptors have been cloned from mammals (there exists one oxytocin and three vasopressin receptors in humans), lower vertebrates, and invertebrates and they are structurally and evolutionarily clearly related to each other (both within a mammalian species and across the different animal classes and phyla) (33–35). The mammalian oxytocin receptors are often involved in various aspects of reproduction (estrous cycle length, partner bond, sexual behavior, birth, milk ejection during lactation, and offspring care) (33). Similar functions of these receptors can be found in other vertebrates and even in invertebrates, such as snails (33–35). The involvement of the oxytocin/vasopressin receptors with reproductive processes, has thus been conserved during a very long period of animal evolution. The obvious question that might be raised, therefore, is in how far insect AKH and mammalian GnRH receptors are functionally related. Does sugar and fat mobilization have something to do with sex and reproduction?
Acknowledgments
We thank Drs. S. Rees and J. Stables (Glaxo Wellcome) for supplying cell line CHO/G16, Lotte Steffensen for typing the manuscript, and Lundbeck Foundation, the Danish Biotechnological Research and Development Program of the Danish Research Agency, and Novo Nordisk Foundation for financial support.
Abbreviations
- AKH
adipokinetic hormone
- BAKHR
Bombyx AKH receptor
- Bom-AKH
Bombyx AKH
- CID
collision induced dissociation
- CHO
Chinese hamster ovary
- Drm-AKH
Drosphila AKH
- DAKHR
Drosophila AKH receptor
- GnRH
gonadotropin-releasing hormone
- GnRHR
GnRH receptor-related
- G16
G protein-16
- Hez-HrTH
Heliothis zea hypertrehalosaemic hormone
- mtAEQ
mitochondrially targeted aequorin
- PCG.6
CHO/G16 cell line stably transfected with the Drosophila AKH receptor DNA
- Scg-AKH
Schistocerca gregaria AKH
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Gäde G, Hoffmann K H, Spring J H. Physiol Rev. 1997;77:963–1032. doi: 10.1152/physrev.1997.77.4.963. [DOI] [PubMed] [Google Scholar]
- 2.Schoofs L, Veelaert D, Vanden Broek J, De Loof A. Peptides. 1997;18:145–156. doi: 10.1016/s0196-9781(96)00236-7. [DOI] [PubMed] [Google Scholar]
- 3.Hewes R S, Taghert P H. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gäde G. Z Naturforsch. 1996;51C:607–617. doi: 10.1515/znc-1996-9-1001. [DOI] [PubMed] [Google Scholar]
- 5.Stone J V, Mordue W, Batley K E, Morris H R. Nature (London) 1976;263:207–211. doi: 10.1038/263207a0. [DOI] [PubMed] [Google Scholar]
- 6.Scarborough R M, Jamieson G C, Kalish F, Kramer S J, McEnroe G A, Miller C A, Schooley D A. Proc Natl Acad Sci USA. 1984;81:5575–5579. doi: 10.1073/pnas.81.17.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaffer M H, Noyes B E, Slaughter C A, Thorne G C, Gaskell S J. Biochem J. 1990;269:315–320. doi: 10.1042/bj2690315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodrik D, Socha R, Simek P, Goldsworthy G J. Insect Biochem Mol Biol. 2000;30:489–498. doi: 10.1016/s0965-1748(00)00025-4. [DOI] [PubMed] [Google Scholar]
- 9.Köllisch G V, Lorenz M W, Kellner R, Verhaert P D, Hoffmann K H. Eur J Biochem. 2000;267:5502–5508. doi: 10.1046/j.1432-1327.2000.01611.x. [DOI] [PubMed] [Google Scholar]
- 10.Siegert K J, Kellner R, Gäde G. Insect Biochem Mol Biol. 2000;30:1061–1067. doi: 10.1016/s0965-1748(00)00081-3. [DOI] [PubMed] [Google Scholar]
- 11.Lorenz M W, Kellner R, Vökl W, Hoffmann K H, Woodring J. J Insect Physiol. 2001;47:563–571. doi: 10.1016/s0022-1910(00)00133-5. [DOI] [PubMed] [Google Scholar]
- 12.Stables J, Green A, Marshall F, Fraser N, Knight E, Sautel M, Milligan G, Lee M, Rees S. Anal Biochem. 1997;252:115–126. doi: 10.1006/abio.1997.2308. [DOI] [PubMed] [Google Scholar]
- 13.Hauser F, Søndergaard L, Grimmelikhuijzen C J P. Biochem Biophys Res Commun. 1998;249:822–828. doi: 10.1006/bbrc.1998.9230. [DOI] [PubMed] [Google Scholar]
- 14.Hauser F, Nothacker H-P, Grimmelikhuijzen C J P. J Biol Chem. 1997;272:1002–1010. doi: 10.1074/jbc.272.2.1002. [DOI] [PubMed] [Google Scholar]
- 15.Wysocki V H, Tsaprailis G, Smith L L, Breci L A. J Mass Spectrom. 2000;35:1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Noyes B E, Katz F N, Schaffer M H. Mol Cell Endocrinol. 1995;109:113–141. doi: 10.1016/0303-7207(95)03492-p. [DOI] [PubMed] [Google Scholar]
- 17.Lenz C, Williamson M, Grimmelikhuijzen C J P. Biochem Biophys Res Commun. 2000;273:1126–1131. doi: 10.1006/bbrc.2000.3062. [DOI] [PubMed] [Google Scholar]
- 18.Veenstra J A. Biochem Biophys Res Commun. 1994;204:292–296. doi: 10.1006/bbrc.1994.2458. [DOI] [PubMed] [Google Scholar]
- 19.Ziegler R, Jasensky R D, Morimoto H. Regul Pept. 1995;57:329–338. doi: 10.1016/0167-0115(95)00046-e. [DOI] [PubMed] [Google Scholar]
- 20.Ziegler R, Cushing A S, Walpole P, Jasensky R D, Morimoto H. Peptides. 1998;19:481–486. doi: 10.1016/s0196-9781(97)00421-x. [DOI] [PubMed] [Google Scholar]
- 21.Vroemen S F, Van der Horst D J, Van Marrewijk W J A. Mol Cell Endocrinol. 1998;141:7–12. doi: 10.1016/s0303-7207(98)00079-3. [DOI] [PubMed] [Google Scholar]
- 22.Velentza A, Spiliou S, Poulos C P, Goldsworthy G J. Peptides. 2000;21:631–637. doi: 10.1016/s0196-9781(00)00200-x. [DOI] [PubMed] [Google Scholar]
- 23.Fernlund P, Josefsson L. Science. 1972;177:173–175. doi: 10.1126/science.177.4044.173. [DOI] [PubMed] [Google Scholar]
- 24.Mordue W, Stone J V. Gen Comp Endocrinol. 1977;33:103–108. doi: 10.1016/0016-6480(77)90132-0. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe H, Raina A K, Riley C T, Fraser B A, Bird T G, Tseng C M, Zang Y S, Hayes D K. Biochem Biophys Res Commun. 1988;155:344–350. doi: 10.1016/s0006-291x(88)81091-x. [DOI] [PubMed] [Google Scholar]
- 26.Ishibashi J, Kataoka H, Nagasawa H, Isogai A, Suzuki A. Biosci Biotech Biochem. 1992;56:66–70. [Google Scholar]
- 27.Birgül N, Weise C, Kreienkamp H-J, Richter D. EMBO J. 1999;18:5892–5900. doi: 10.1093/emboj/18.21.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenz C, Søndergaard L, Grimmelikhuijzen C J P. Biochem Biophys Res Commun. 2000;269:91–96. doi: 10.1006/bbrc.2000.2251. [DOI] [PubMed] [Google Scholar]
- 29.Lenz C, Williamson M, Grimmelikhuijzen C J P. Biochem Biophys Res Commun. 2000;273:571–577. doi: 10.1006/bbrc.2000.2964. [DOI] [PubMed] [Google Scholar]
- 30.Lenz C, Williamson M, Hansen G N, Grimmelikhuijzen C J P. Biochem Biophys Res Commun. 2001;286:1117–1122. doi: 10.1006/bbrc.2001.5475. [DOI] [PubMed] [Google Scholar]
- 31.Auerswald L, Birgül N, Gäde G, Kreienkamp H-J, Richter D. Biochem Biophys Res Com. 2001;282:904–909. doi: 10.1006/bbrc.2001.4659. [DOI] [PubMed] [Google Scholar]
- 32.Secher T, Lenz C, Cazzamali G, Sørensen G, Williamson M, Hansen G N, Svane P, Grimmelikhuijzen C J P. J Biol Chem. 2001;276:47052–47060. doi: 10.1074/jbc.M106675200. [DOI] [PubMed] [Google Scholar]
- 33.Gimpl G, Fahrenholz F. Physiol Rev. 2001;81:629–683. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 34.Van Kesteren R E, Smit A B, De Lange R P, Kits K S, Van Golen F A, Van der Schors R C, De With N D, Burke J F, Geraerts W P M. J Neurosci. 1995;15:5989–5998. doi: 10.1523/JNEUROSCI.15-09-05989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Kesteren R E, Tensen C P, Smit A B, Van Minnen J, Kolakowski L F, Meyerhof W, Richter D, Van Heerikhuizen H, Vreugdenhil E, Geraerts W P M. J Biol Chem. 1996;271:3619–3626. doi: 10.1074/jbc.271.7.3619. [DOI] [PubMed] [Google Scholar]