Abstract
The histidine containing phospho carrier protein (HPr) kinase/phosphatase is involved in carbon catabolite repression, mainly in Gram-positive bacteria. It is a bifunctional enzyme that phosphorylates Ser-46-HPr in an ATP-dependent reaction and dephosphorylates P-Ser-46-HPr. X-ray analysis of the full-length crystalline enzyme from Staphylococcus xylosus at a resolution of 1.95 Å shows the enzyme to consist of two clearly separated domains that are assembled in a hexameric structure resembling a three-bladed propeller. The N-terminal domain has a βαβ fold similar to a segment from enzyme I of the sugar phosphotransferase system and to the uridyl-binding portion of MurF; it is structurally organized in three dimeric modules exposed to form the propeller blades. Two unexpected phosphate ions associated with highly conserved residues were found in the N-terminal dimeric interface. The C-terminal kinase domain is similar to that of the Lactobacillus casei enzyme and is assembled in six copies to form the compact central hub of the propeller. Beyond previously reported similarity with adenylate kinase, we suggest evolutionary relationship with phosphoenolpyruvate carboxykinase. In addition to a phosphate ion in the phosphate-binding loop of the kinase domain, we have identified a second phosphate-binding site that, by comparison with adenylate kinases, we believe accommodates a product/substrate phosphate, normally covalently linked to Ser-46 of HPr. Thus, we propose that our structure represents a product/substrate mimic of the kinase/phosphatase reaction.
Keywords: signaling‖catabolite repression‖P-loop‖PEPCK‖AdK
The bacterial phosphoenolpyruvate (PEP)-dependent sugar phosphotransferase system (PTS) is a multicomponent transport system responsible for the uptake of carbohydrates. By using PEP as a primary phosphoryl group donor it catalyzes the phosphotransfer via the two general components enzyme I (EI) and histidine phospho carrier protein HPr to sugar specific enzyme II (EII) complex composed of hydrophilic domains EIIA and EIIB and the membrane-spanning domain EIIC (1). Besides its function as a group translocation system for carbohydrate uptake, PTS is a signaling device; it is linked to nitrogen metabolism (2), chemotaxis toward the sugar substrate (3), and additional regulatory functions (4).
In enteric bacteria, the regulation of carbohydrate metabolism is mediated by EIIAglc of the PTS, which is the soluble part of the glucose-sensing membrane-associated EII complex (5, 6), whereas in Gram-positive bacteria, the regulation centers on the phospho-carrier protein HPr. HPr contains two phosphorylation sites. During the PEP-dependent phospho transfer, HPr is phosphorylated at His-15 by EI. In addition, HPr is subject to regulatory phosphorylation at Ser-46 by the ATP:Mg2+-dependent HPr kinase/phosphatase (HPrK), a general sensor for ATP, free phosphate (Pi), and glycolytic intermediates (7–10). The bifunctional enzyme has been identified in several Gram-positive microorganisms (7, 11–13) and some pathogenic bacteria (14–16) as well. ATP-dependent protein kinases have long been known to play a key role in the regulation of metabolism and signal transduction in eukaryotic organisms (17) but were discovered later also in prokaryotes (18, 19). The lack of sequence similarities of HPr kinase/phosphatase to other protein kinases indicates that HPrK represents a new family of bacterial ATP-dependent protein kinases.
In the presence of glucose and certain rapidly metabolizable carbohydrates, HPrK preferentially acts as a kinase, stimulated by high ATP levels. In addition, a stimulatory effect of fructose-1,6-bisphosphate (F-1,6-BP) can be observed, which differs among the organisms previously described. In Bacillus subtilis, the HPrK is active only in the presence of F-1,6-BP (11), whereas in Streptococcus salivarius, Enterococcus faecalis and Mycoplasma pneumoniae this metabolite does not affect the kinase activity of the enzyme (7, 12, J. Stülke, personal communication). For HPrK of Staphylococcus xylosus, it could be observed that F-1,6-BP indeed increases the kinase activity in the presence of 2 mM ATP, albeit to a lower extent. Possibly, other unidentified effectors may exist that are dependent on the external energy and carbon availability and, therefore, of the natural habitat of each organism.
Phosphorylation of HPr on Ser-46 induces various effects on carbohydrate metabolism. First, PEP-dependent phosphorylation of HPr is diminished drastically and, in consequence, sugar uptake via PTS is strongly decreased (20). As an additional function, P-Ser-46-HPr serves as a signaling molecule and acts as a transcriptional corepressor in Gram-positive bacteria. It is involved in carbon catabolite repression and activation through its interaction with catabolite control protein A (21–23), a transcription factor of the GalR/LacI family of regulatory proteins (24, 25).
To understand the mechanistic principles of HPrK activity at the atomic level, we have crystallized the full-length HPrK from Staphylococcus xylosus and determined its structure at 1.95 Å resolution by x-ray crystallography. Our results will be discussed in light of biochemical and structural work on related systems.
Materials and Methods
Protein Expression and Purification.
The hpr K/P gene from S. xylosus was derived from plasmid pKIN7 (13) and amplified by PCR. The PCR product (950 bp) containing the gene was cloned blunt end in the SmaI site of pUC20 (Roche Molecular Biochemicals) and sequenced. Subsequently, the fragment was excised from this vector with NdeI and BamHI and cloned into pET11a (Novagen) in which the gene could be expressed under the control of the T7 promoter in the strain E. coli BL21 (DE 3; Stratagene). Cells (11.2 g) were harvested by centrifugation, resuspended in 20 ml of buffer A (50 mM Tris⋅HCl, pH 7.5/10−4 M PMSF/10−4 M EDTA/10−4 M NaN3/10−4 M DTT) and disrupted by ultrasonic treatment. Cell debris was removed by centrifugation for 15 min at 20 000 × g; the supernatant containing membrane fragments was centrifuged twice for 1.5 h at 150 000 × g, dialyzed against buffer A for at least 12 h, and applied to a Q-Sepharose column (3 cm × 10 cm; Amersham Pharmacia). After gradient elution, the pool was adjusted to 20% ammonium sulfate and loaded on a Butyl TSK column (5 cm × 20 cm; TosoHaas, Montgomeryville, PA) preequillibrated with 20% ammonium sulfate in buffer A. HPrK eluted at 8.8–13.2% ammonium sulfate. The pool was concentrated and loaded onto a Sephadex G-200 gel filtration column (5 cm × 90 cm; Amersham Pharmacia). The HPrK pool was desalted on a Sephadex G-25 column (2 cm × 22,5 cm; Amersham Pharmacia) equilibrated with 50 mM ammonium hydrogen carbonate. After lyophilization, the pool contained 130–140 mg homogeneous HPrK. For crystallization experiments, the protein was dissolved in 50 mM Hepes, pH 7.5 and concentrated to about 40 mg/ml by using a centricon concentrator (Amicon).
Crystallization and X-Ray Analysis.
Rhombohedral crystals of space group R32 were grown at room temperature (≈20°C) from hanging drops composed of equal volumes (typically 2 + 2 μl) of protein (10–20 mg/ml) and crystallization buffer (1.6 M Na/KPi, pH 7,6; Hampton Research, Riverside, CA), suspended over a reservoir containing 500 μl of crystallization buffer. The crystals appeared after 2–5 days and grew to a full size of typically 200 × 200 × 100 μm3 within ≈2 weeks. The presence of 0.5 mM AppNHp in the crystallization medium seemed to improve crystal size. For data collection, crystals were transferred to reservoir solution containing 15% ethylene glycol as cryoprotectant and were subsequently frozen in a cryogenic nitrogen stream of 100 K temperature. Initial data were recorded at beamlines ID13 and ID14–2 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. A data set of 1.95 Å resolution was collected from one single crystal at beam line ID14–4 at ESRF. Data processing and scaling were done with the program XDS (26).
The structure was determined with the molecular replacement method using the coordinates of the C-terminal domain of HPrK from L. casei (27) as a search model in calculations with the program CNS (28). Strong tetrahedral maxima of electron density within the P-loop region and in the subunit interface were interpreted as phosphate ions, based on the crystallization from Na/KPi. After several rounds of model building and refinement, all done with O and CNS, respectively, and the addition of solvent molecules, we arrived at a model that consists of two chains comprising almost all residues (excluding 233–245 and 299–314) of HPrK, three phosphate moieties each, and 293 solvent molecules. Visual representation of the structural models was done with the programs MOLSCRIPT (29) and RASTER3D (30).
Results and Discussion
Structure Determination and Overall Architecture.
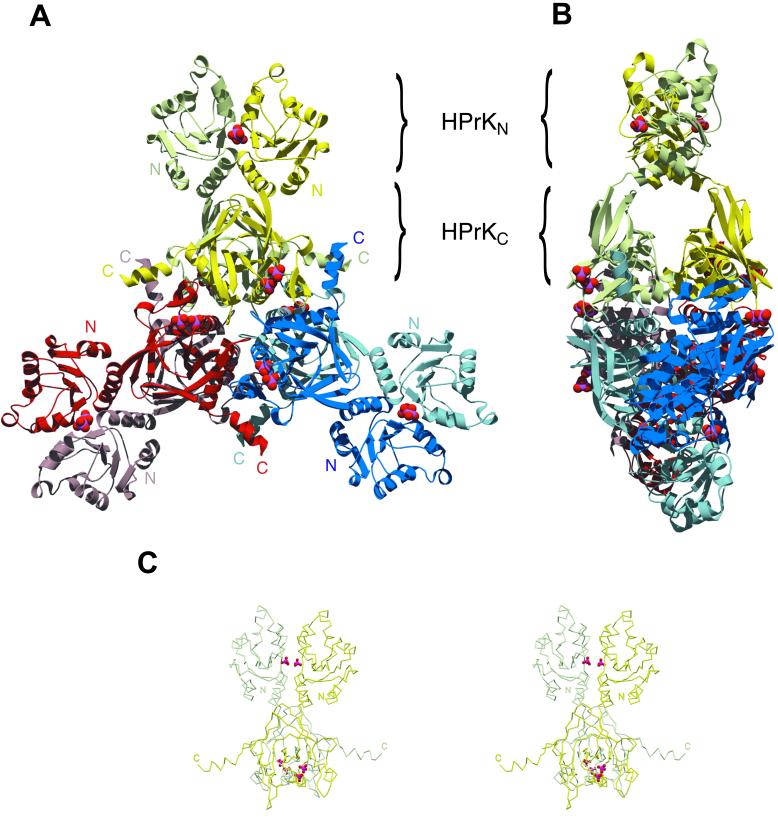
Full-length HPrK (314 amino acids) from S. xylosus was overexpressed in Escherichia coli, purified, and crystallized as described in Methods. The structure was solved with the molecular replacement method, using the structure of the C-terminal (kinase) domain of Lactobacillus casei (27) as a search model. The current model of the asymmetric unit, refined to a resolution of 1.95 Å with Rwork and Rfree of 22.6% and 24.6%, respectively (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org), includes two chains of HPrK and contains all residues except for 233–245 and 299–314, which are disordered in the crystal. The crystallographic analysis is summarized in Table 1. The segment 233–245 is in an exposed area and is cleaved in limited proteolysis experiments (S. Hasenbein, unpublished observations). Three neighboring dimers in the crystal lattice are strongly intertwined, giving rise to a compact hexameric structure. The overall shape resembles a three-bladed propeller of 150 Å in diameter and 60 Å in thickness. The blades are formed by two N-terminal domains each, and the compact central hub assembles the C-terminal kinase domains (Fig. 1). Analytical ultracentrifugation experiments (data not shown) support the idea that, like the L. casei enzyme (27) the functional HPrK from S. xylosus is indeed a hexamer, giving an estimated molecular mass of 223.5 kDa, which is in reasonable agreement with the expected value of 212 kDa (6 × 35.3 kDa) for a hexameric HPrK. The asymmetric distribution of intermolecular contacts gives the impression of a hexamer built from two trimers.
Figure 1.
Structure of the full-length HPr protein kinase from Staphylococcus xylosus. (A) Front view of the HPrK hexamer, composed of three structurally identical dimers deeply intertwined. The kinase domains (HPrKC) occupy the central part of the structure whereas the N-terminal domains (HPrKN) extend outwards as pairwise blades in a propeller. Phosphate ions in the interface between N-terminal domains and P-loop regions are depicted as CPK models. (B) Side view of the hexamer. (C) Stereo view of the HPrK dimer representing the asymmetric unit of the crystal.
Architecture of the HPrK Subunit.
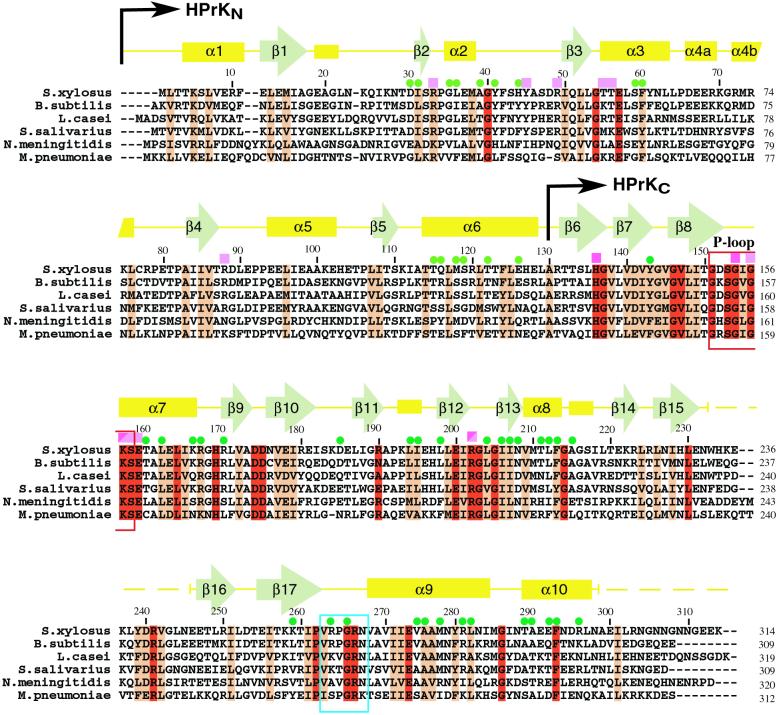
The structural model of a single subunit reveals two clearly separated domains: an N-terminal mixed βαβ-like domain (HPrKN, residues 1–130) and a C-terminal domain (HPrKC, residues 131–298) responsible for kinase/phosphatase activity (Fig. 1; ref. 27). Fig. 2 shows a sequence alignment of various HPrK species with the assignment of secondary structure elements for our model included. HPrKC is very similar to the corresponding domain of HPrK from L. casei, and according to the program DALI (31), resembles adenylate kinase (AdK)-like proteins (27, 32) and is related to the C-terminal domain of PEP carboxykinase (PCK, see below) (34, 35).
Figure 2.
Multiple sequence alignment of representative members of the HPrK protein family with the secondary structure assignment included (Top), as derived by the program DSSP (47). N-terminal (HPrKN) and C-terminal (HPrKC) domains are indicated. Dashed lines show sequence regions that are not ordered in the crystals (see text). The P-loop region is indicated as a red box. Pink squares denote residues that interact with phosphate ions, and green dots indicate amino acids involved in inter-subunit interactions in the N-terminal (light green) and C-terminal (dark green) domains. In HPrKC, light-pink squares indicate typical P-loop phosphate interactions, and dark-pink squares mark interactions with the second phosphate molecule (see text). Residues K157 and R202 contact both phosphates. Based on structural similarities to NMP kinases and PCK, the region composed of amino acids 263 to 268 (cyan box) is predicted to interact with the adenine moiety of the ATP.
The two domains are connected by an α-helix (α6) and do not show apparent intra-molecular contacts. The two chains in the asymmetric unit are arranged in a parallel orientation: the two N-terminal domains from different chains (see below) interact with each other to form a dimeric structure (Fig. 3) as do the two C-terminal domains. In this parallel orientation, HPrKN and HPrKC dimers do not show apparent crossdomain contacts with each other within the asymmetric unit, forming clearly distinct structural elements (Fig. 1C).
Figure 3.
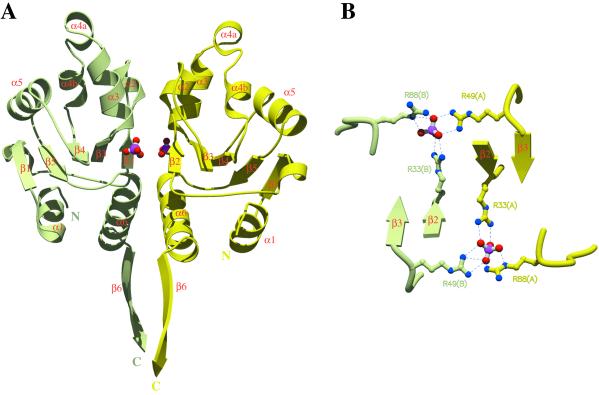
HPrKN and phosphate binding. (A) Ribbon diagram of the dimeric HPrKN, colored as in Fig. 1. The first strand of the C-terminal domain (β6) has been included to illustrate the relative orientation of N-terminal and C-terminal domains (see also Fig. 1C). The two phosphate ions, represented in ball and stick, are stabilized by interactions with residues at the subunit interface. Details of these interactions are displayed in B and Fig. 5. (B) Six arginine residues, contributed by both subunits, interact with the phosphate ions to form an almost planar network (bottom view with respect to A).
Domain HprKN, a Phosphate-Binding Module?
Our structure analysis reveals that the previously uncharacterized N-terminal segment (1–130) folds as an independent globular domain (HPrKN). It consists of a central five-stranded mixed β-sheet surrounded by eight helices (Figs. 2 and 3). A search with DALI (31) indicated significant similarity (score 4.2) with the archaeal thermosome (35) and with the α/β subdomain from EI of the PTS system (36, 37), but not with its HPr binding domain (ref. 38, see Note 1, which is published as supporting information on the PNAS web site). The dimer interface between two HPrKN partners is made up of almost exclusively polar interactions involving mostly conserved residues (Figs. 2 and 3) and a number of water molecules.
A striking and unexpected feature in the HPrKN dimer interface is the presence of two tetrahedrally shaped electron-density peaks located at the N-termini of the helix dipoles derived from helices α3 and α6. Given the crystallization from sodium/potassium phosphate, we interpret them as Pi ions (39). Beyond interactions with the main chain amides derived from α3, they are stabilized by intra- (mediated by Arg 33/88, Thr-55/56/114) and inter-subunit (mediated by Arg-49, Tyr-45) contacts (Fig. 3 and Fig. 5, which is published as supporting information on the PNAS web site). The distance between the two phosphorus atoms is about 17 Å. The structural arrangement of the phosphates along with stabilizing arginines resembles a planar network in the interface between the two N-terminal domains (Fig. 3).
The function associated with the N-terminal domain is presently not well characterized at the biochemical level. If it contributes to enzyme activity, it is likely to have only a regulatory role, because the C-terminal domain from L. casei has a catalytic activity similar to the full-length enzyme (27). From our structure, it is likely that binding of a phosphorylated compound/protein is playing a role in its function. Indeed, a number of phosphate-containing substrates have been shown to affect HPrK activity. However, current biochemical evidence supports the view that regulation by Pi and probably also by F-1,6-BP does not require the N-terminal domain. The structure does not suggest how regulatory signals would be transmitted from the phosphate-binding site in HPrKN to the active site of HPrKC (Fig. 1C, see below). Given its relative structural independence, it is possible that HPrKN represents a module of a distinct function, for example, a protein–protein interaction domain. As already mentioned by Brochu and Vadeboncoeur (7), glycolyzing cells of Streptococcus salivarius also contain P-His-15-HPr and doubly phosphorylated (P-Ser-46)(P-His-15)HPR, which could influence the HPrK activity. Also, phosphorylated EIIA- or EIIB-domains of the glucose specific enzyme II complex, which are present in starving Gram-positive bacterial cells, may exhibit regulatory functions mediated by the N-terminal domain. Site-directed mutagenesis of the phosphate-binding region of HPrKN would be helpful to characterize its contribution to HPrK function. Additional structures of the full-length enzyme in complex with substrates and effectors would be required to give further insight into the regulation of HPrK.
Domain HPrKC, a Product/Substrate Mimic?
HPrKC is very similar to the corresponding domain from the L. casei enzyme (27). The two molecules have an amino acid sequence identity of 48% and superimpose with an rmsd value of 1.3 Å, aligning 148 Cα atoms out of 155 corresponding residues. It is made up of a central five-stranded mixed β-sheet flanked by four α-helices and by two smaller β-sheets (Fig. 6, which is published as supporting information on the PNAS web site). According to DALI (31), the structure resembles AdK (27, 32) and has an even higher similarity with the C-terminal domain of PCK (Fig. 6; ref. 33), which DALI returns with a score of 7.8, aligning 128 structurally equivalent Cα atoms with an rmsd value of 2.8 Å. A multiple sequence alignment (not shown), including members of HPrK and PCK families, indicates significant sequence similarity, suggesting that the two proteins are, in fact, evolutionarily related.
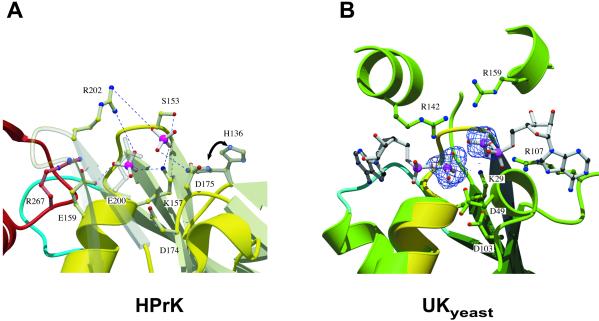
It was not unexpected from the crystallization conditions to find a phosphate ion Pi in the P-loop (ref. 40; residues 151–158) that normally accommodates the β-phosphate of the bound nucleotide, similar to what was observed previously in HPrKC from L. casei or earlier in adenylate kinase (41). It is stabilized in the known way by the helix dipole of α7, main chain amides, sharing typical Pi contacts with the conserved Lys-157 of the phosphate binding (P) loop motif (GxxxxGKS/T; ref. 40). Additional contributions come from Arg-202 and Arg-267 of a neighboring subunit contacting the Pi by means of a water molecule (Fig. 6).
In addition to the P-loop-bound phosphate, we have identified a second phosphate-binding site, not observed in the L. casei HPrK structure (27), in close proximity. The distance between the two phosphorus atoms in our structure is about 4.5 Å. The second phosphate is stabilized by polar interactions with the highly conserved Ser-153, Lys-157, and Arg-202 (Fig. 2 and Fig. 4). In trying to understand whether this site represents aspects of the biochemical reactions catalyzed by HPrK or whether it is just a favorable binding site, we compared our model with PCK (34) and different AdK-related NMP kinases in substrate and product-bound states (32). Strikingly, in a superposition of uridylate kinase from yeast (UKyeast) bound to two ADP molecules (42, 43), the β-phosphate positions of the bound nucleotides almost perfectly match the sites occupied by the two phosphate moieties in our structure (Fig. 4). Therefore, we reasoned that, by analogy, the second phosphate in our HPrKC domain might in fact represent the position of the γ-phosphate of ATP after its transfer to Ser-46 of HPr. To test this hypothesis, we superimposed the phosphate group of p-Ser-46-HPr from E. faecalis (44) onto the refined position of the second phosphate in HPrKC. This alignment resulted in a docking model that would require only moderate conformational changes to form a sterically favorable complex. In addition, the environment of the phosphate moiety and of the docked HPr is surrounded by a number of invariant residues, including His-136, Asp-174/Asp-175, Ser-153, Glu-200, and by Phe-293 from a neighboring subunit. In this scenario, as in the docking model obtained previously by simulated annealing (27), the flexible loop (233–245) containing additional highly conserved residues might become ordered upon interaction with HPr. A similar comparison with the more closely related PCK will have to await a structural model of a product-bound state of this enzyme.
Figure 4.
HPrKC and phosphate binding. (A) Two phosphate ions are bound in the P-loop region, stabilized by invariant residues, most likely involved in catalysis (H136, K157, Ser-153, Glu-159, R202, E200) and/or metal binding (Asp-174/175) (see text). Phosphate 1 occupies the position of the β-phosphate of ATP and is additionally stabilized by water-mediated contacts with R267 from a neighboring subunit. Phosphate 2 (closest to S153) we propose to represent the position of the γ-phosphate of ATP after its transfer to Ser-46 of HPr. Major interactions stabilizing the phosphate ions are shown as blue dashed lines. H136 seems to adopt alternative conformations in the structure, both of which are displayed. (B) P-loop region of HPrKC (yellow) superimposed on the AdK-related UKyeast, bound to two ADP molecules (PDB code 1uky), shown in ball and stick. Electron density corresponding to the two phosphate ions found in HPrKC has been included, matching nearly perfectly the β-phosphate positions of ADP bound to UKyeast. Residues in UKyeast that are structurally equivalent to those in the P-loop of HPrKC are also displayed. K29 and R142 of UKyeast, equivalent to K157 and R202, contact both phosphate ions and are believed to play a role in catalysis through stabilization of transition states. The putative adenine-binding loops are depicted in cyan; the corresponding loop (blue) from neighboring subunit of HPrK is included.
The analysis of the phosphate binding area in HPrKC has revealed remarkable similarities with the active sites of UKyeast and other AdK-related NMP kinases (Fig. 4) and even more with PCK, although some residues are derived from topologically different regions. Residues contacting the phosphate moieties seem to be functionally homologous to their counterparts in UKyeast or PCK. For example, Lys-157 and Arg-202 contact both phosphate moieties in a conformation very similar to that seen in UKyeast and other NMP kinases and are likely to be involved in catalysis (45). Indeed, mutation of Lys-157 to methionine in HPrK from E. faecalis abolishes kinase and phosphatase activities (46). Likewise, mutations of residues in the phosphate-binding area of HPrK from Mycoplasma pneumoniae, including counterparts of Arg-202, Ser-153, and Glu-159, differentially affect kinase and phosphatase activities (Stülke, personal communication). It seems that with the basic structural arrangement found in HPrK, PCK, and NMP kinases, nature has evolved a protein scaffold suitable for phosphotransfer reactions on multiple substrates, ranging from nucleotides and metabolic intermediates to high-molecular weight substrates like proteins.
Conclusions
The bifunctional HPrK protein kinase/phosphatase plays a key role in carbon catabolite repression. High ATP levels and low Pi concentrations, concomitant with high nutritional states, stimulate the kinase activity. In contrast, low ATP and high Pi concentrations, occurring during starvation, inhibit the kinase and stimulate the phosphatase activity. Our structural model revealed the modular architecture of the protein assembled in a propeller-shaped hexamer and showed features of the catalytic machinery in great detail. The structural model sets the stage for further functional analysis of the protein; in particular, the role of the previously uncharacterized N-terminal domain as a putative regulatory device is now accessible to targeted investigation of relevant structural/functional features that may be involved in the regulation of the balance between both activities. Compared with the C-terminal catalytic region, the N-terminal domain is less conserved among the HPrKs from various microorganisms and thus may reflect a species-specific adaptation to varying environmental conditions. The common features of the catalytic machinery in HPrK, PCK, and AdK-related NMP kinases suggests that nature has recruited similar structural scaffolds to catalyze phosphotransfer to different types of substrates.
Supplementary Material
Acknowledgments
We thank Reinhold Brückner, University of Kaiserslautern for providing the plasmid pKIN7 containing the hprK gene of S. xylosus; the staff at the beam lines ID13, ID14–1/4 of the European Synchrotron Facility, Grenoble, France for technical support; K. Steinhauer and J. Stülke for providing their manuscript describing mutational analyses of HPrK of M. pneumoniae before publication. J.A.M. gratefully acknowledges support from the Ramón Areces Foundation (Madrid, Spain). S.H., B.K., and W.H. were supported by Deutsche Forschungsgemeinschaft, SFB 394. We are indebted to Matti Saraste who died during the completion of this work; we deeply miss him, his scientific support, and his encouragement.
Abbreviations
- PEP
phosphoenolpyruvate
- PTS
phosphotransferase system
- EI
enzyme I
- EII
enzyme II
- HPrK
HPr kinase/phosphatase
- AdK
adenylate kinase
- PCK
PEP carboxykinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1ko7).
References
- 1.Postma P W, Lengeler J W, Jacobson G R. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reizer J, Reizer A, Saier M H, Jr, Jacobson G R. Protein Sci. 1992;1:722–726. doi: 10.1002/pro.5560010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengeler J W, Vogler A P. FEMS Microbiol Rev. 1989;5:81–92. doi: 10.1016/0168-6445(89)90011-9. [DOI] [PubMed] [Google Scholar]
- 4.Gunnewijk M G, van den Bogaard P T, Veenhoff L M, Heuberger E H, de Vos W M, Kleerebezem M, Kuipers O P, Poolman B. J Mol Microbiol Biotechnol. 2001;3:401–413. [PubMed] [Google Scholar]
- 5.Inada T, Kimata K, Aiba H. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 6.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. Proc Natl Acad Sci USA. 1997;94:12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brochu D, Vadeboncoeur C. J Bacteriol. 1999;181:709–717. doi: 10.1128/jb.181.3.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutscher J, Saier M H., Jr Proc Natl Acad Sci USA. 1983;80:6790–6794. doi: 10.1073/pnas.80.22.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reizer J, Novotny M J, Hengstenberg W, Saier M H., Jr J Bacteriol. 1984;160:333–340. doi: 10.1128/jb.160.1.333-340.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reizer J, Sutrina S L, Saier M H, Stewart G C, Peterkofsky A, Reddy P. EMBO J. 1989;8:2111–2120. doi: 10.1002/j.1460-2075.1989.tb03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galinier A, Kravanja M, Engelmann R, Hengstenberg W, Kilhoffer M C, Deutscher J, Haiech J. Proc Natl Acad Sci USA. 1998;95:1823–1828. doi: 10.1073/pnas.95.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kravanja M, Engelmann R, Dossonnet V, Bluggel M, Meyer H E, Frank R, Galinier A, Deutscher J, Schnell N, Hengstenberg W. Mol Microbiol. 1999;31:59–66. doi: 10.1046/j.1365-2958.1999.01146.x. [DOI] [PubMed] [Google Scholar]
- 13.Huynh P L, Jankovic I, Schnell N F, Bruckner R. J Bacteriol. 2000;182:1895–1902. doi: 10.1128/jb.182.7.1895-1902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 15.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, et al. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 17.Hunter T. Methods Enzymol. 1991;200:3–37. doi: 10.1016/0076-6879(91)00125-g. [DOI] [PubMed] [Google Scholar]
- 18.LaPorte D C. J Cell Biochem. 1993;51:14–18. doi: 10.1002/jcb.240510104. [DOI] [PubMed] [Google Scholar]
- 19.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stulke J, Karamata D, Saier M H, Jr, Hillen W. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 20.Monedero V, Poncet S, Mijakovic I, Fieulaine S, Dossonnet V, Martin-Verstraete I, Nessler S, Deutscher J. EMBO J. 2001;20:3928–3937. doi: 10.1093/emboj/20.15.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deutscher J, Kuster E, Bergstedt U, Charrier V, Hillen W. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 22.Jones B E, Dossonnet V, Kuster E, Hillen W, Deutscher J, Klevit R E. J Biol Chem. 1997;272:26530–26535. doi: 10.1074/jbc.272.42.26530. [DOI] [PubMed] [Google Scholar]
- 23.Turinsky A J, Grundy F J, Kim J H, Chambliss G H, Henkin T M. J Bacteriol. 1998;180:5961–5967. doi: 10.1128/jb.180.22.5961-5967.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 25.Weickert M J, Adhya S. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 26.Kabsch W. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 27.Fieulaine S, Morera S, Poncet S, Monedero V, Gueguen-Chaignon V, Galinier A, Janin J, Deutscher J, Nessler S. EMBO J. 2001;20:3917–3927. doi: 10.1093/emboj/20.15.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 30.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 31.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 32.Vonrhein C, Schlauderer G J, Schulz G E. Structure (London) 1995;3:483–490. doi: 10.1016/s0969-2126(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 33.Matte A, Goldie H, Sweet R M, Delbaere L T. J Mol Biol. 1996;256:126–143. doi: 10.1006/jmbi.1996.0072. [DOI] [PubMed] [Google Scholar]
- 34.Matte A, Tari L W, Goldie H, Delbaere L T. J Biol Chem. 1997;272:8105–8108. doi: 10.1074/jbc.272.13.8105. [DOI] [PubMed] [Google Scholar]
- 35.Ditzel L, Lowe J, Stock D, Stetter K O, Huber H, Huber R, Steinbacher S. Cell. 1998;93:125–138. doi: 10.1016/s0092-8674(00)81152-6. [DOI] [PubMed] [Google Scholar]
- 36.Liao D I, Silverton E, Seok Y J, Lee B R, Peterkofsky A, Davies D R. Structure (London) 1996;4:861–872. doi: 10.1016/s0969-2126(96)00092-5. [DOI] [PubMed] [Google Scholar]
- 37.Garrett D S, Seok Y J, Liao D I, Peterkofsky A, Gronenborn A M, Clore G M. Biochemistry. 1997;36:2517–2530. doi: 10.1021/bi962924y. [DOI] [PubMed] [Google Scholar]
- 38.Garrett D S, Seok Y J, Peterkofsky A, Gronenborn A M, Clore G M. Nat Struct Biol. 1999;6:166–173. doi: 10.1038/5854. [DOI] [PubMed] [Google Scholar]
- 39.Copley R R, Barton G J. J Mol Biol. 1994;242:321–329. doi: 10.1006/jmbi.1994.1583. [DOI] [PubMed] [Google Scholar]
- 40.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 41.Dreusicke D, Schulz G E. FEBS Lett. 1986;208:301–304. doi: 10.1016/0014-5793(86)81037-7. [DOI] [PubMed] [Google Scholar]
- 42.Muller-Dieckmann H J, Schulz G E. J Mol Biol. 1994;236:361–367. doi: 10.1006/jmbi.1994.1140. [DOI] [PubMed] [Google Scholar]
- 43.Muller-Dieckmann H J, Schulz G E. J Mol Biol. 1995;246:522–530. doi: 10.1006/jmbi.1994.0104. [DOI] [PubMed] [Google Scholar]
- 44.Audette G F, Engelmann R, Hengstenberg W, Deutscher J, Hayakawa K, Quail J W, Delbaere L T. J Mol Biol. 2000;303:545–553. doi: 10.1006/jmbi.2000.4166. [DOI] [PubMed] [Google Scholar]
- 45.Tsai M D, Yan H G. Biochemistry. 1991;30:6806–6818. doi: 10.1021/bi00242a002. [DOI] [PubMed] [Google Scholar]
- 46.Kravanja M. Dissertation. Bochum, Germany: Ruhr-Universität; 1999. [Google Scholar]
- 47.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.