Abstract
We have defined inactive α and ω fragments of β-lactamase that can complement to form a functional enzyme in both bacteria and mammalian cells, serving as a readout for the interaction of proteins fused to the fragments. Critical to this advance was the identification of a tripeptide, Asn-Gly-Arg, which when juxtaposed at the carboxyl terminus of the α fragment increased complemented enzyme activity by up to 4 orders of magnitude. β-Lactamase is well suited to monitoring constitutive and inducible protein interactions because it is small (29 kDa), monomeric, and assayable with a fluorescent cell-permeable substrate. The negligible background, the magnitude of induced signal caused by enzymatic amplification, and detection of signal within minutes are unparalleled in mammalian protein interaction detection systems published to date.
Protein–protein interactions are involved in every cellular process ranging from gene expression and signal transduction to cell division and differentiation, yet they have been among the most difficult aspects of cell biology to study. Standard biochemical methods have yielded most of the available information about such interactions, but these assays often are limited by the available reagents such as monoclonal antibodies for immunoprecipitation or lack the appropriate cellular context.
The development of fusion protein-based assays such as the yeast two-hybrid method (1) has expanded the potential for studying protein interactions in intact cells greatly. However, this assay relies on the transcription of a reporter gene; consequently it is not applicable to studies of the kinetics of protein–protein interactions and is unable to detect the interaction of compartmentalized proteins such as receptors at the cell surface. A method based on fluorescence resonance energy transfer provided a further advance and currently is one of the most accurate methods used to monitor dynamic interactions (2). However, the incremental changes in fluorescence assayed by fluorescence resonance energy transfer are small, and the stringent steric requirements for detecting the interacting proteins can restrict the utility of this technique.
Assays based on the complementation of enzyme fragments fused to interacting proteins that regenerate enzymatic activity after dimerization are particularly well suited for monitoring inducible protein interactions (reviewed in ref. 3). These systems have important advantages including low-level expression of the test proteins, generation of signal as a direct result of the interaction, and enzymatic amplification. As a result, they are highly sensitive and physiologically relevant assays (4). Additionally, assays based on enzyme complementation can be performed in any cell type of interest or in diverse cellular compartments such as the nucleus, secretory vesicles, or plasma membrane.
The class A β-lactamases are particularly attractive candidates for an assay based on enzyme fragment complementation because of the fact that they are monomeric and of relatively small size (5). In addition, β-lactamases have been expressed successfully in prokaryotic and eukaryotic cells, making this system applicable to both classes of organisms (6). We identified a pair of β-lactamase fragments (α197 and ω198) that complemented to produce detectable activity in bacteria when fused to two helices that form a leucine zipper (7). Detectable interactions were not limited to these moieties, because interactions between larger proteins were detected also.
To increase the signal-to-noise ratio associated with the β-lactamase assay, we screened libraries of random tripeptides inserted at the break-point termini of the α197 and ω198 fragments for peptides that enhanced complementation of the enzyme. We report the identification of a tripeptide, Asn-Gly-Arg (NGR), that produced a profound enhancement of βlactamase activity mediated by different protein pairs in bacteria when introduced at the carboxyl terminus of the α197 fragment.
We reasoned that extension of the β-lactamase system into mammalian cells would provide significant advantages over other fragment complementation systems currently used [e.g., βgalactosidase (8) and dihydrofolate reductase (9), because the fragments are small (<19 kDa)], there is no endogenous β-lactamase activity, and a highly sensitive cell-permeable fluorescent substrate has been developed recently (10). The β-lactamase fragments were tested to determine whether they could be used to monitor inducible interactions in a mammalian cell line measured either by fluorescence microscopy or flow cytometry. Here we show that the β-lactamase fragments also could detect inducible interactions in eukaryotic cells. Finally, the observed β-lactamase complementation was a direct measure of enzyme activity, not dependent on de novo protein synthesis, and generated detectable signal within minutes of protein dimerization, making it applicable to the detection of transient protein interactions. This system should have broad utility in monitoring protein interactions in diverse intracellular compartments in a range of cell types.
Materials and Methods
Construction and Screening of the Tripeptide Libraries in Escherichia coli.
The β-lactamase α197 fragment was amplified first from pUC19 by PCR with forward primer JH101 (5′-CTGTGCCATGGTGAGTATTCAACATTTCCGTGTCG-3′) and reverse degenerate primer JH303 (5′-CTAGGAATTCMYBMYBMYBTTCGCCAGTTAATAGTTTGC-3′). The PCR product was subcloned into a p15A-based bacterial expression vector as a fusion to the carboxyl terminus of the Jun helix. TG1 cells were transformed and plated on 2xYT plates, and 1 × 104 independent colonies were collected after overnight growth at 33°C. The ω198 library was constructed similarly with forward degenerate primer JH302 (5′-GCATGGTACCVRKVRKVRKCTACTTACTCTAGCTTCCCG-3′) and reverse primer JH16 (5′-CCGGAATTCTTACCAATGCTTAATCAGTGAGGC-3′). The PCR product was subcloned into a pUC-based phagemid vector between the KpnI and EcoRI sites as a fusion protein to the amino terminus of the Fos helix. The library was transformed, and ≈1 × 104 independent colonies were isolated. The phagemid library was rescued with a helper phage, R408 (Stratagene), and used to infect TG1 cells expressing the α197-Jun construct including the peptide library. Infected cells (1 × 108) were plated on increasing concentrations of ampicillin (50, 100, 200, 400, and 800 μg/ml). After an overnight incubation, colonies appearing on plates containing the highest ampicillin concentrations were collected. To separate the two vectors expressing the α197 and ω198 fragments, DNA isolated from each putative clone was used to transform DH5α cells and then selected on plates containing either kanamycin or chloramphenicol for isolation of each vector expressing either the α197-peptide-Jun or the Fos-peptide-ω198 fusion protein, respectively. Colonies resistant to one antibiotic but not the other were isolated.
β-Lactamase Activity Assay in E. coli.
Cells in 2xYT medium with 10% glucose and appropriate antibiotics were grown to a density of 0.3 OD600 at 33°C and then induced with 0.1 mM isopropyl β-D-thiogalactoside for 2 h without antibiotics. The culture (750 μl) was incubated with 1.5 ml of 0.1 mg/ml nitrocefin. The absorbance of 200 μl of cell-free supernatant was measured at 490 nm in a BioRad Benchmark microplate reader (Bio-Rad).
Detection of β-Lactamase Fragments in E. coli by Immunoblot.
Cells in 2xYT medium containing 2% Glucose, 34 mg/ml chloramphenicol, and 35 mg/ml kanamycin were induced at an OD600 of 0.4 with 0.1 mM isopropyl β-D-thiogalactoside for 3 h at 33°C. Cells (4 × 108) were lysed directly into SDS sample buffer or by probe sonication in PBS supplemented with 150 mM NaCl/1% Triton X-100/5% Glycerol/10 mM benzamidine/Sigma E. coli-specific protease inhibitor mixture/1 mM PMSF and partitioned into soluble and insoluble fractions by centrifugation. Lysates were subjected to SDS/PAGE on a 4–20% gradient gel. Proteins were Western-blotted with horseradish peroxidase-conjugated anti-Flag monoclonal antibody (Sigma) and detected by using the ECL+Plus detection kit (Amersham Pharmacia).
Retroviral Vectors for Expression of β-Lactamase Fragments in Mammalian Cells.
To create the β-lactamase fusion proteins for retroviral expression in mammalian cells, an oligonucleotide encoding a GS linker, 5′-TCGAGGGTGGAGGCGGTTCAGGCGGAGGTGGCAGCGGCGGTGGCGGATCGG, was inserted into the XhoI/SalI site of both pWZL-neo and pWZL-hygro. The α197-NGR fragment was amplified by PCR from plasmid FHT 4002A1 by using the primers 5′-CTCGAGCACCCAGAAACGCTGG and 3′-GTCGACTTCCCGCCCATTTTCG. The ω198 fragment was amplified by using the primers 5′-CTCGAGGGAGTGCAGGTGGAAACC and 3′-CTCGACTTCCAGTTTTAGAAGC. The α197 fragment was cloned into the XhoI site of pWZL-GS-Neo, and the ω198 fragment was cloned into the SalI site of pWZL-GS-Hygro. The FKBP-rapamycin binding domain (FRB) of FKBP-rapamycin-associated protein corresponding to amino acid residues 2,025–2,114 of human FRAP was cloned as an XhoI/SalI fragment into the SalI site of pWZL-α197-GS-Neo. The full-length coding sequence of FKBP12 was cloned as a SalI/XhoI fragment into the XhoI site of pWZL-GS-ω198-hygro. The extracellular and transmembrane regions of epidermal growth factor receptor corresponding to amino acids 1–655 (4) was cloned as an NcoI/BamHI fragment into the pWZL-FKBP12ω198-Hygro vector. The wild-type β-lactamase was expressed from a pWZL vector also encoding puromycin resistance, which was a generous gift from Garry Nolan (Stanford University School of Medicine, Stanford, CA).
Retroviral Production, Infection, and Mammalian Cell Culture.
The ecotropic ΦNX-packaging cell line (P. L. Achacoso and G. P. Nolan, unpublished data) was transiently transfected with the proviral constructs by using fugene transfection reagent (Roche Molecular Biochemicals). The supernatant from the transfected cells was removed 48–72 h later and applied to C2C12 myoblasts. Polybrene was added to a final concentration of 8 μg/ml (Sigma). Transduced cells were selected and maintained in the appropriate antibiotic (hygromycin or neomycin, Invitrogen) at a concentration of 1 mg/ml. C2C12 myoblasts were grown in DMEM (Invitrogen)/20% FBS. Cells were treated with 50 nM rapamycin unless otherwise stated.
β-Lactamase Assayed by CCF2/AM Staining, Immunofluorescence, and Fluorescence-Activated Cell Sorter (FACS) Analysis in Mammalian Cells.
Immunofluorescence.
To assay β-lactamase in C2C12 myoblasts, the CCF2/AM substrate (Aurora Biosciences, San Diego) was used at a final concentration of 2 μM in DMEM with 2.5 mM probenecid. Cells were washed once in PBS and then incubated for 30 min with the CCF2/AM substrate at a concentration of 3 × 105 cells per ml. The plate was washed three times in PBS and visualized with a β-lactamase filter set (Chroma Technology, Brattleboro, VT; excitation 405 ± 10 nm, 425 dichroic mirror, 435-nm long-pass emission).
FACS.
Data were collected on a modified FACStar plus (Becton Dickinson) with Moflo electronics (Cytomation, Fort Collins, CO). Cells were trypsinized, washed twice in PBS, incubated with CCF2/AM substrate (as described above) for 1 h, and then washed twice in a PBS/5% FBS solution. Ten thousand events were collected for each sample.
Results
Peptide Selection in E. coli.
Chimeric proteins were constructed by using the complementing α197 and ω198 β-lactamase fragments and the leucine zipper helices from the c-Fos (Fos) and c-Jun (Jun) subunits of the AP-1 transcription factor (11). Because the Fos and Jun helices interact constitutively, they provided a system that could be used to screen libraries for peptides that enhanced interaction-dependent enzyme complementation. The leucine zipper helices were fused at the break-point termini of the β-lactamase fragments via flexible (Gly4Ser)3 linkers. The α197 β-lactamase fragment was fused to the amino terminus of the Fos helix and coexpressed with the ω198 fragment fused to the carboxyl terminus of the Jun helix (Fig. 1A). Synthetic oligonucleotides encoding random tripeptides were inserted into the coding sequence of each fragment between the break-point and the flexible (Gly4Ser)3 linker that connects the interacting helices to each fragment (Fig. 1B). The library was designed to encode only three amino acids to avoid selecting high-affinity peptides, which themselves would mediate dimerization of the fragments. The libraries also were enriched for charged amino acids, because charge–charge interactions were likely to have the greatest effect. This enrichment was achieved by using the degenerate codon VRK (cag/ag/tg) that can only encode His, Gln, Arg, Asn, Lys, Ser, Asp, Glu, and Gly.
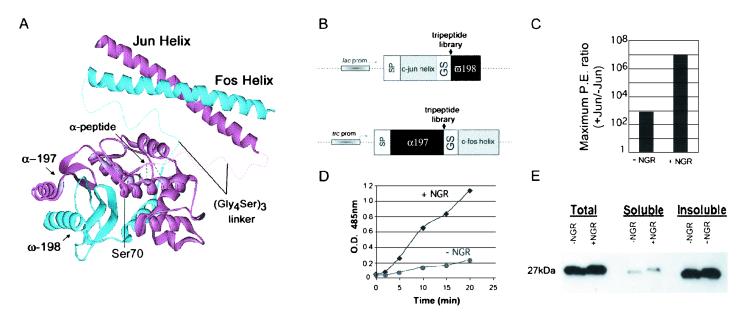
Figure 1.
β-Lactamase fragment structure and complementation assays in bacteria. (A) Structure of TEM-1 β-lactamase and illustrations of its reconstitution from the α197 (amino acids 25–197) and ω198 (amino acids 198–288) fragments connected by the flexible linker (Gly4Ser)3 and brought into contact by dimerization of the Fos and Jun helices. Placement of the α-tripeptide library is labeled with possible contacts as well as the active site serine (Ser-70). (B) Vectors used for expression of β-lactamase fragment fusion proteins with random tripeptide libraries inserted at the fragmentation point. Also indicated are signal peptide (SP), lactose operon promoter (lac prom), fusion of the tryptophan operon, lactose operon promoters (trc prom), and the glycine-serine linker (GS) as described for A. (C) Cells expressing the complementing constructs, α197Jun and ω198Fos, were plated on a range of ampicillin concentrations (10–200 μg/ml). The number of colonies obtained (signal) was divided by the number of colonies derived from plating the same constructs without the Jun helix (background) on the corresponding ampicillin concentration. The same assay was performed with the α197 fragment containing the NGR peptide insertion. The largest ratio obtained for each was designated as the maximum plating efficiency (P.E.) ratio, which is representative of five independent experiments. (D) Total β-lactamase activity was determined by assaying cells expressing the α197Jun and ω198Fos with and without the NGR insertion. The cells were induced to express the fusion proteins and assayed at equivalent cell densities in excess substrate (nitrocefin). Absorbance at 485 nM was determined in the cell-free supernatants of each sample and plotted against time. (E) To determine the effect of the NGR peptide on the stability of the β-lactamase α197 fragment, cells (from D) at 2 h of induction were lysed, and the accumulated protein was immunoblotted by using an antibody to the flag-tagged α197 fragment.
The α197-Jun and Fos-ω198 constructs containing the peptide library insertions were cotransformed into E. coli cells. The cells were plated on solid medium containing a concentration of ampicillin that allowed only 0.001% of cells expressing the complementing parent fusion constructs to survive and produce colonies. Bacterial resistance to the antibiotic ampicillin is conferred through the activity of β-lactamase; therefore colony growth in the presence of ampicillin is indicative of β-lactamase activity. All colonies underwent two cycles of recovery, dilution, and replating at 10–100 cells per colony to ensure that only clones with plating efficiencies above 1% (or 1,000-fold higher than the parent) were retained. Increased resistance to ampicillin was indicative of improved β-lactamase complementation.
Twelve clones exhibiting enhanced ampicillin resistance were recovered in this manner and examined further. In all cases only the α fragment tripeptide enhanced β-lactamase activity, and no selected ω-fragment tripeptide had a significant effect either with or without its associated α-tripeptide. Four different αtripeptides were selected: Gly-Arg-Glu (GRE), NGR, His-Ser-Glu (HSE), and Glu-Lys-Arg (EKR). Of these, the NGR peptide had the greatest effect and was chosen for further study.
Effect of Selected Peptides on Signal-to-Noise Ratio.
As an indication of the selectivity of the system for interacting proteins relative to the background resulting from coexpression of the fragments, the maximum ratio of interaction-dependent activity to interaction-independent activity was determined and designated as the plating-efficiency ratio (Fig. 1C). Interaction-dependent activity was assessed by plating cells coexpressing the α197-Jun fusion containing the NGR peptide and the Fos-ω198 fusion on concentrations of ampicillin ranging from 10 to 200 μg/ml. The background activity, or interaction-independent activity, was measured in the same manner except that the Jun helix was removed from the α197-NGR construct. Cells expressing the parental constructs without the selected NGR peptide exhibited a 1,000-fold higher survival rate than cells in which the Jun helix had been removed, resulting in a maximum plating-efficiency ratio of 1 × 103. By contrast, the NGR peptide exhibited a maximum plating-efficiency ratio of 1 × 107. Thus, the mere presence of the NGR tripeptide between the break-point of the α197 fragment and the linker enabled a 1,000,000-fold enrichment of the Fos–Jun interaction over proteins lacking Jun (no interaction) in a single plating compared with only a 1,000-fold enrichment for the same constructs lacking NGR.
Although the increase in plating efficiency observed for the NGR peptide was encouraging, the possibility remained that the effect was specific for the Fos–Jun interaction. Previously we used the β-lactamase α197/ω198 fragment complementation system to isolate proteins that bound specifically to the extracellular domain of the human immune cell coactivation antigen CD40 [CD40ED (12)] from a library of random 12-mer peptides expressed within the context of the thioredoxin protein (ref. 13; J.-H.H. and R.B., unpublished results). The peptide showing the highest affinity for CD40ED, BW10-1, was used to test the effect of the NGR peptide on complementation mediated by this interaction.
The NGR peptide produced substantial increases in the complementation because of the interaction of BW10-1 with CD40ED over a range of ampicillin concentrations, resulting in a maximum plating-efficiency ratio that was greater than 12-fold higher than the interaction in the absence of the peptide (Table 1). Thus, although the effects of the NGR peptide on the CD40–thioredoxin protein interactions were less than on the Fos–Jun helix interaction, the enhanced ampicillin resistance and signal-to-noise ratio demonstrate that the effect of the NGR peptide is not limited to the Fos–Jun interaction.
Table 1.
Complementation of α197-CD40ED and BW10-1-ω198 in E. coli
| Ampicillin Concentration, μg/ml
|
|||
|---|---|---|---|
| 10 | 25 | 50 | |
| +NGR, +CD40/−CD40, % | 100/4.00 | 100/0.08 | 100/<0.01 |
| Ratio | 25 | 1,250 | >10,000 |
| −NGR, +CD40/−CD40, % | 100/0.12 | 19/0.07 | 1.1/<0.01 |
| Ratio | 833 | 271 | >110 |
α197 fused to BW10-1 with or without the NGR peptide was coexpressed with ω198 with or without CD40ED. Ten thousand cells expressing both constructs were plated on increasing concentrations of ampicillin, and the number of colonies formed are expressed as a percentage of cells plated. These data are representative of three independent experiments. <0.01 indicates that no colonies appeared when 104 cells were plated. Maximum plating-efficiency ratios are in boldface and were determined as described for Fig. 1C.
Effect of Selected Tripeptides on β-Lactamase Enzymatic Activity.
To ensure that the increases in ampicillin resistance conferred by the NGR peptide were caused by increases in β-lactamase activity, we measured total enzyme activity with the chromogenic substrate nitrocefin (14). Cells expressing the complementing fusion proteins, Fos-ω198 and α197-Jun, with and without the NGR peptide were grown in suspension cultures and resuspended at the same cell density in isotonic buffer containing an excess of the substrate. The substrate and product readily diffuse into and out of the periplasm enabling the time-dependent accumulation of colored product in the cell-free supernatant (Fig. 1D). This assay also showed that the NGR peptide conferred a large increase in β-lactamase activity relative to controls without peptide, confirming that the observed increases in plating efficiency correlated with β-lactamase enzyme activity.
The increase in β-lactamase activity observed in the presence of the NGR tripeptide could be caused by stabilization by NGR of the α197 fragment and consequently the complemented enzyme. To determine whether addition of the NGR peptide led to accumulation of the α fragment, the amount of α197-Jun was determined by immunoblot when coexpressed with the complementing Fos-ω198 with and without NGR (Fig. 1E). The NGR peptide had no discernible effect on the abundance of soluble α-fragment fusion protein in either the total cell lysate or the soluble fraction, indicating that the mechanism by which NGR increases β-lactamase activity does not involve stabilizing the α197 chimera as assayed by Western blot.
β-Lactamase Complementation in Mammalian Cells.
Experiments were designed to test whether the β-lactamase fragments in conjunction with the NGR peptide could be used to monitor an inducible protein interaction in mammalian cells (Fig. 2). The well characterized inducible interaction of FKBP12 and FRB was used as a model system (15–19). FKBP12 (FK506-binding protein 12) binds FRB only in the presence of the pharmacological agent rapamycin. Rapamycin is a small cell-permeable molecule that can be added directly to the culture medium resulting in heterodimerization of FKBP12 and FRB. Two fusion proteins were constructed: by using flexible linkers (Gly4Ser)3, FKBP12 was fused to the amino terminus of the ω198 fragment, and FRB fused to the carboxyl terminus of the α197 fragment containing the NGR peptide (Fig. 2A). The bacterial signal sequence from each of the β-lactamase fusion fragments was removed. The fusion constructs were expressed by using pWZL retroviral vectors that encode proteins conferring resistance to hygromycin or neomycin. The pWZL vectors were selected for use because they are expressed at relatively low levels; in these vectors the splice donor/acceptor is deleted, resulting in reduced translation efficiency in mammalian cells compared with other retroviral vectors such as MFG (8, 20). Thus, these vectors avoid vast overexpression of proteins and more closely approximate physiological levels.
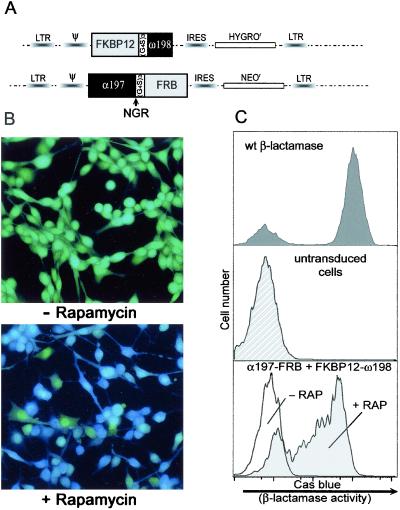
Figure 2.
Inducible β-lactamase fragment complementation in C2C12 myoblasts. (A) Schematic of the bicistronic, retrovirally expressed fusion proteins FKBP12ω198 and α197FRB with selectable markers for hygromycin (HYGROR) and neomycin (NEOR) driven by an internal ribosome entry site (IRES). Ψ designates the viral packaging signal and LTR marks the long terminal repeats. (B) Immunofluorescence assay of β-lactamase activity. C2C12 cells expressing the FKBP12ω198 and α197FRB fusions were loaded with the cell-permeable CCF2/AM substrate in the absence (Upper, −) and presence (Lower, +) of rapamycin (2 h) and then imaged by fluorescence microscopy. Green indicates intact substrate, and blue indicates cleaved substrate. (C) FACS analysis of β-lactamase activity. Cells with and without rapamycin treatment (2 h) were trypsinized, loaded with CCF2/AM substrate, and assayed by flow cytometry. Increases in cascade blue fluorescence indicate β-lactamase activity (log scale). (Top) β-Lactamase staining of cells expressing wild-type β-lactamase. (Middle) Untransduced cells stained with the CCF2 substrate. (Bottom) Cells expressing the β-lactamase fusion constructs with and without rapamycin. Dimerization of the fusion constructs induced by rapamycin causes a 50–100-fold increase in cascade blue fluorescence from the responding population.
A stable cell line containing the FKBP12ω198-hygro and α197FRB-neo constructs was established through retroviral infection of C2C12 cells and subsequent antibiotic selection. Cells from this population were treated with 50 nM rapamycin for 2 h and assayed for β-lactamase activity by using the fluorogenic CCF2/AM substrate (10). The intact CCF2/AM substrate, when excited by a UV wavelength of 409 nm, emits at 520 nm (green), whereas after cleavage by β-lactamase it emits at 447 nm (blue). As shown in Fig. 2B (Top), the cells expressing the fusion proteins appear green in the absence of rapamycin, indicating that little or no cleavage of the substrate has occurred. However, after exposure to rapamycin the substrate is cleaved, shifting the fluorescence from green to blue, indicating reconstitution of β-lactamase activity (Fig. 2B, Bottom). These results revealed that inducible dimerization of FKBP12 and FRB could lead to the complementation of the β-lactamase fragments, resulting in functional β-lactamase activity in mammalian cells.
These data were confirmed by performing a quantitative measurement of β-lactamase activity by flow cytometry (FACS) using the CCF2/AM substrate (Fig. 2C). The histograms of the cells that stably expressed FKBP12ω198 and α197FRB in the absence of rapamycin (Bottom) overlapped with and were not significantly different from untransduced negative control cells (Middle). By contrast, after exposure of the cells harboring the β-lactamase fragments to rapamycin for 2 h, enzyme activity was induced substantially, and an increase in fluorescence of 50–100-fold above background was evident. Two features of these data are particularly noteworthy: (i) the almost undetectable background activity resulting from complementation in the absence of rapamycin and (ii) the marked increase (orders of magnitude) in the signal generated by the complementation.
Surprisingly, 20% of the cells expressing wild-type βlactamase did not stain positive for β-lactamase activity even though the cells were kept in continuous drug selection to ensure retention of the virus containing the wild-type β-lactamase gene. A similar percentage of nonresponding cells (≈23%) can be seen in the population of cells expressing the chimeric β-lactamase proteins in the presence of rapamycin either by flow cytometry or fluorescence imaging. This phenomenon was noted also in the original study describing the CCF2/AM substrate with similar ratios (80% responding and 20% not responding), suggesting that it may be a feature of the substrate-staining procedure itself (10). Thus 80% seems to be the maximum number of cells that can be expected to stain positive by using this assay.
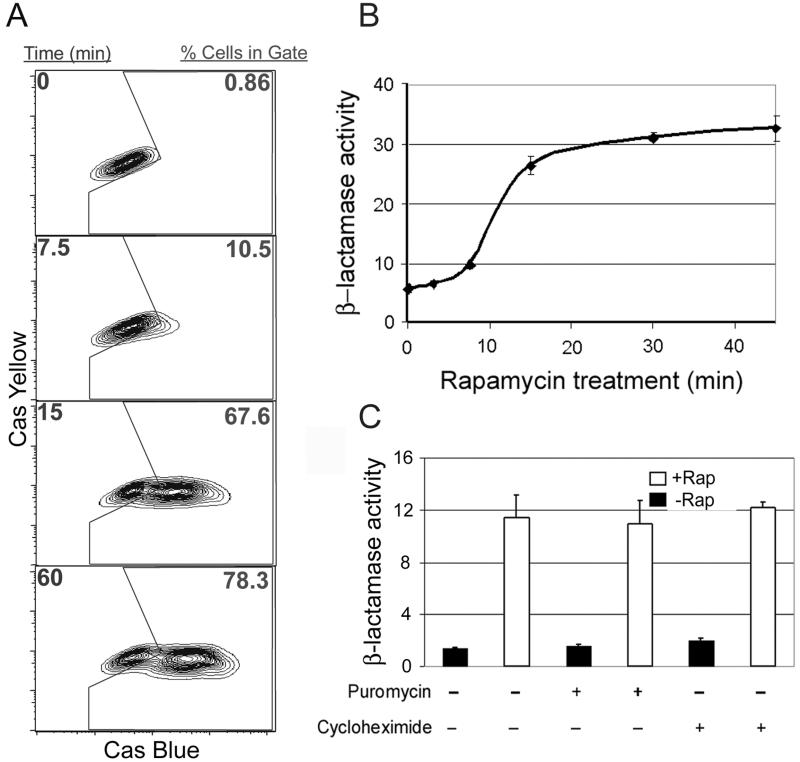
Fig. 3A shows a time course of rapamycin treatment in C2C12 cells expressing the chimeric β-lactamase fusion proteins that demonstrated the ability to distinguish quantitatively the responding from the nonresponding population. When a gate was drawn around the cells expressing the fusion constructs before induction (time 0), it only included 0.9% of the cells. Notably, 10% of the cells were positive at 7.5 min after rapamycin addition, and at 1 h 76% of the population stained positive for β-lactamase activity, i.e., most if not all of the cells capable of responding. Longer rapamycin treatment did not increase the numbers of positive cells significantly.
Figure 3.
Time course of inducible β-lactamase complementation in C2C12 myoblasts. (A) Time course of rapamycin-induced dimerization. The FKBP12ω198-α197FRB cell line was assayed for β-lactamase activity by FACS. Cells were stained with the CCF2/AM substrate, treated with rapamycin, and assayed over time. The gate is represented in the center of each plot, and the percentage of cells falling within this region are shown in red. (B) Mean fluorescence time course. FACS data (from A) represented as mean cascade (Cas) blue fluorescence were calculated in triplicate and graphed over time. (C) Inhibition of protein synthesis does not affect rapamycin-induced complementation. α197FRB-FKBP12ω198 cells were treated with either puromycin or cycloheximide (100 μg/ml) for 2 h before the addition of rapamycin (1 h). The cells were stained with the CCF2/AM substrate and assayed by flow cytometry. The mean fluorescence for the cascade blue channel was calculated in triplicate and graphed on the y axis.
The time course was rapid and began to plateau within 15 min. This result is most clearly evident when the data from a FACS analysis performed in triplicate are presented as mean fluorescence (Fig. 3B). After rapamycin addition, a response was seen as early as 7.5 min after rapamycin treatment and was 70% maximal within 15 min, demonstrating that by using a bulk assay for fluorescence, the generated signal is detectable also within minutes of induced complementation. These kinetics are significantly faster than those reported for other systems using the FKBP12-FRB proteins to induce dimerization (21, 22), which demonstrates the high specific activity of the complemented enzyme and the extreme sensitivity of the system.
The rapid kinetics of β-lactamase reconstitution after the addition of rapamycin suggested that de novo protein synthesis might not be necessary. To test this possibility, we assayed β-lactamase activity in the presence of the protein synthesis inhibitors puromycin and cycloheximide at concentrations of 100 μg/ml for 2 h before the addition of rapamycin. Neither of these inhibitors significantly altered the amount of complementation observed relative to the controls, indicating that de novo protein synthesis is not necessary for β-lactamase complementation (Fig. 3C). Many inducible protein–protein interactions have been documented to occur on a time scale of seconds to minutes. The data shown here suggest that the β-lactamase system has the potential to monitor not only rapid but possibly also transient protein–protein interactions.
Detection of Constrained Protein–Protein Interactions in Mammalian Cells.
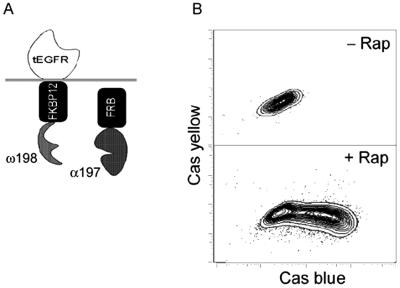
Cell surface-mediated signal transduction events often require the interaction of a membrane-associated protein such as a transmembrane receptor with a cytoplasmic protein. To test whether the β-lactamase system is capable of monitoring interactions in such a constrained configuration, we constructed a model membrane-bound protein that would interact with a cytoplasmic protein only in the presence of an inducer. For this purpose, a tripartite fusion construct comprised of the extracellular and transmembrane region of epidermal growth factor receptor, which anchored the protein to the plasma membrane fused to the FKBP12ω198 chimera (Fig. 4), was generated (4). This fusion protein was coexpressed with a cytoplasmic α197-FRB chimera as described previously. Cells expressing these constructs were assayed by FACS for induction of β-lactamase activity in the presence of rapamycin (Fig. 4B). The magnitude of β-lactamase complementation observed when proteins are in a constrained membrane-anchored conformation is comparable to that observed when the proteins are expressed freely in the cytoplasm (compare Fig. 3A with Fig. 4B).
Figure 4.
Interaction of a membrane-bound and cytoplasmic protein assayed by β-lactamase complementation in C2C12 myoblasts. (A) Diagram depicting the membrane-bound and cytoplasmic fusion proteins that were coexpressed in C2C12 cells. The truncated epidermal growth factor receptor (tEGFR) was used to tether the FKBP12ω198 to the plasma membrane. (B) Cells expressing the fusion constructs in the absence (Upper) or presence (Lower) of rapamycin (1 h) were assayed by flow cytometry. Cas, cascade.
Discussion
Protein fragment complementation assays have widespread potential for understanding biological processes, because they can be adapted to high-throughput assays, cDNA screens, and the study of inducible protein interactions. Such biosensors of protein–protein interactions should be invaluable in elucidating signal transduction pathways in specific cells (transformed, differentiated, and dividing) in response to well defined extracellular stimuli such as hormones, cytokines, and calcium. Moreover, they can be used to screen for molecules that promote or disrupt such interactions, which could serve not only as invaluable biological tools but also be applied to drug discovery.
Although several systems have been developed that use chimeras of proteins of interest and enzyme fragments to assess protein interactions, each has its limitations. For example, in mammalian cells the fluorescent signal generated by complementation of dihydrofolate reductase is not amplified enzymatically; thus, only small increments in fluorescence are achieved (9, 23). The βgalactosidase system benefits from enzymatic amplification of its signal; however, the active enzyme is a homotetramer, and the individual fragments are large (80 kDa), making it likely that some interactions may be sterically hindered (4, 8). The small size, monomeric nature, and availability of a cell-permeable fluorescent substrate suggest that a system based on the β-lactamase enzyme has the potential to overcome many of the limitations of existing systems. Indeed, the β-lactamase complementation system described here exhibits an extremely high signal-to-noise ratio measured by plating efficiency in bacteria or in mammalian cells by flow cytometry and fluorescence imaging. In addition, the ability to generate a signal within minutes and the capacity to perform the assay in the absence of de novo protein synthesis suggests that this system may be ideal for studying inducible and transient protein–protein interactions in any cell type.
We have shown in bacteria that by screening an oligonucleotide library a tripeptide sequence could be isolated that greatly enhanced the complementation of the α197 and ω198 pair of βlactamase fragments. The NGR peptide was shown to be effective in a leucine zipper helix interaction as well as a more complex CD40–thioredoxin protein interaction, increasing the overall sensitivity of the system by up to 3 orders of magnitude in the colony formation assay based on growth in the presence of ampicillin. Although this peptide had a direct effect on reconstituted βlactamase activity in bacteria, the increase in activity was not caused by an increase in the stability of the α fragment or accumulation of the complemented enzyme assayed by Western blot. This finding suggests that the NGR peptide may act, at least in part, by increasing the specific activity of the reconstituted enzyme. This hypothesis is supported by analysis of three-dimensional structures for TEM-1 β-lactamase (24). We examined potential interactions between side chains of the NGR peptide and residues underlying the fragmentation site in the reconstituted enzyme. This modeling suggested that the NGR peptide could make five different contacts on three distinct helices, including two contacts on helix 2. Coincidentally, helix 2 contains the active site nucleophile Ser-70. Thus, we speculate that interactions with the helix 2 residues could stabilize the active site, increasing the specific activity of the complemented enzyme, accounting for the apparent increase in enzymatic activity.
The magnitude of complementation observed for the βlactamase system in the presence of the NGR peptide led us to test the applicability of the system in assaying protein interactions in mammalian cells. By using the inducible FKBP12-FRB dimerization system, we showed that it is not only possible to monitor an inducible interaction in mammalian cells by using β-lactamase complementation but also that this assay yields a very robust signal of 50–100-fold increase in fluorescence from the responding cell population. This finding, as well as the negligible background observed from the expression of the fusion proteins in the absence of a dimerizing agent, makes the highly sensitive measurement of protein interactions using this system readily apparent.
Properties inherent to the β-lactamase system suggest that it approaches a physiologically relevant measure of protein interactions in mammalian cells. The α197 fragment is ≈19 kDa, whereas the ω198 fragment is only ≈10 kDa. These values are smaller than many proteins used to monitor protein localization such as green fluorescent protein, making it unlikely that the fragments will alter the function of the chimeric proteins being analyzed significantly. The assay can be performed in any cell type and can be used to assay dimerization irrespective of protein localization. In addition, we have shown that we can detect interactions in as little as 7.5 min, and that this activity can occur in the absence of de novo protein synthesis, demonstrating its utility in the study of inducible or transient protein interactions. In summary, we have developed a broadly applicable protein–protein interaction biosensor that has significant advantages over traditional biochemical as well as existing protein fragment complementation systems. This system should enable the identification of molecules that promote or inhibit key protein interactions via high-throughput screens in a range of cell types, phyla, and species. Moreover, given its unique properties, β-lactamase may be particularly well suited to identifying novel protein interactions specific to subcellular compartments of transformed, proliferating, and differentiating cell types via a mammalian two-hybrid assay.
Acknowledgments
We thank Julia Spiegel for invaluable technical assistance and Dr. Fabio Rossi for insightful discussions throughout this research and helpful comments on the manuscript. T.S.W. was supported sequentially by the Genentech-Stanford fellowship, National Institutes of Health (NIH) Aging Training Grant AG00259 and NIH Biotechnology Training Grant GM08142. H.M.B. gratefully acknowledges support from NIH Grants HD18179, AG09521, HL65572, and CA59717, Department of the Army Contract DAMD17-00-1-0442, and Department of Energy Subcontract 6507640.
Abbreviations
- FRB
FKBP-rapamycin binding domain of FKBP-rapamycin-associated protein
- FACS
fluorescence-activated cell sorter
References
- 1.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 2.Adams S R, Harootunian A T, Buechler Y J, Taylor S S, Tsien R Y. Nature (London) 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 3.Rossi F M, Blakely B T, Blau H M. Trends Cell Biol. 2000;10:119–122. doi: 10.1016/s0962-8924(99)01707-9. [DOI] [PubMed] [Google Scholar]
- 4.Blakely B T, Rossi F M, Tillotson B, Palmer M, Estelles A, Blau H M. Nat Biotechnol. 2000;18:218–222. doi: 10.1038/72686. [DOI] [PubMed] [Google Scholar]
- 5.Philippon A, Dusart J, Joris B, Frere J M. Cell Mol Life Sci. 1998;54:341–346. doi: 10.1007/s000180050161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore J T, Davis S T, Dev I K. Anal Biochem. 1997;247:203–209. doi: 10.1006/abio.1997.2092. [DOI] [PubMed] [Google Scholar]
- 7. Balint R, Her, J.-H. (1999) U.S. Patent Application Serial No. 09/526,106.
- 8.Rossi F, Charlton C A, Blau H M. Proc Natl Acad Sci USA. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Remy I, Michnick S W. Proc Natl Acad Sci USA. 1999;96:5394–5399. doi: 10.1073/pnas.96.10.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zlokarnik G, Negulescu P A, Knapp T E, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien R Y. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 11.Karin M, Liu Z, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 12.Durie F H, Foy T M, Masters S R, Laman J D, Noelle R J. Immunol Today. 1994;15:406–411. doi: 10.1016/0167-5699(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 13.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 14.McManus-Munoz S, Crowder M W. Biochemistry. 1999;38:1547–1553. doi: 10.1021/bi9826512. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Zheng X F, Brown E J, Schreiber S L. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. Nature (London) 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 17.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Nature (London) 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 18.Belshaw P J, Ho S N, Crabtree G R, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J, Chen J, Schreiber S L, Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 20.Riviere I, Brose K, Mulligan R C. Proc Natl Acad Sci USA. 1995;92:6733–6737. doi: 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otto K G, Jin L, Spencer D M, Blau C A. Blood. 2001;97:3662–3664. doi: 10.1182/blood.v97.11.3662. [DOI] [PubMed] [Google Scholar]
- 22.Muthuswamy S K, Gilman M, Brugge J S. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remy I, Wilson I A, Michnick S W. Science. 1999;283:990–993. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 24.Jelsch C, Mourey L, Masson J M, Samama J P. Proteins. 1993;16:364–383. doi: 10.1002/prot.340160406. [DOI] [PubMed] [Google Scholar]