Abstract
Formation of mixed disulfides between glutathione and the cysteines of some proteins (glutathionylation) has been suggested as a mechanism through which protein functions can be regulated by the redox status. The aim of this study was to identify the proteins of T cell blasts that undergo glutathionylation under oxidative stress. To this purpose, we radiolabeled cellular glutathione with 35S, exposed T cells to oxidants (diamide or hydrogen peroxide), and performed nonreducing, two-dimensional electrophoresis followed by detection of labeled proteins by phosphorimaging and their identification by mass spectrometry techniques. We detected several proteins previously not recognized to be glutathionylated, including cytoskeletal proteins (vimentin, myosin, tropomyosin, cofilin, profilin, and the already known actin), enzymes (enolase, aldolase, 6-phosphogluconolactonase, adenylate kinase, ubiquitin-conjugating enzyme, phosphoglycerate kinase, triosephosphate isomerase, and pyrophosphatase), redox enzymes (peroxiredoxin 1, protein disulfide isomerase, and cytochrome c oxidase), cyclophilin, stress proteins (HSP70 and HSP60), nucleophosmin, transgelin, galectin, and fatty acid binding protein. Based on the presence of several protein isoforms in control cells, we suggest that enolase and cyclophilin are heavily glutathionylated under basal conditions. We studied the effect of glutathionylation on some of the enzymes identified in the present study and found that some of them (enolase and 6-phosphogluconolactonase) are inhibited by glutathionylation, whereas the enzymatic activity of cyclophilin (peptidylprolyl isomerase) is not. These findings suggest that protein glutathionylation might be a common mechanism for the global regulation of protein functions.
Formation of mixed disulfides between glutathione and cysteines in proteins (glutathionylation) has long been known to occur after oxidative stress (1). Recently, protein glutathionylation has gained attention as a possible means of redox regulation of protein functions.
Glutathionylation takes place by thiol-disulfide exchange between protein sulfhydryls and oxidized glutathione (GSSG), a reversible process that can be catalyzed by glutaredoxin. GSSG/reduced glutathione (GSH) ratio is an indicator of the redox status of the cell, and the extent of protein glutathionylation will vary accordingly: a higher ratio will promote glutathionylation/binding, a lower ratio will result in deglutathionylation/release of glutathione. This finding also suggests a likely compartmentalization of protein glutathionylation, as GSSG/GSH ratio is very high in the endoplasmic reticulum, where an oxidizing environment favors sulfhydryl oxidation (2). Glutathionylated proteins also can be formed by reaction with S-nitrosoglutathione, a pathway requiring the formation of this compound from nitric oxide (3).
A number of proteins have been reported to undergo glutathionylation, including enzymes, transcription factors, and oncogenes (reviewed in ref. 4). The identification of glutathionylated proteins is hampered by the lack of specific antibodies to modified cysteines that would allow detecting glutathionylated proteins by immunoblotting, in analogy with the protocols for the study of phosphorylated proteins.
Typically, proteome analysis using two-dimensional gel electrophoresis followed by MS fingerprinting/sequencing can be used to identify modified proteins. A similar approach has been used to identify carbonyl-modified proteins after oxidative stress in yeast after one-dimensional PAGE (5) and, using two-dimensional gels, to identify vitellogenin as a major oxidized protein in aging Caenorhabditis elegans (6). More recently this technique was applied to the analysis of Alzheimer's disease brains and led to the identification of β-actin and creatine kinase as the two major carbonylated proteins. Finally, a technique was set up to detect S-nitrosylated proteins in neuronal cells and led to the finding of several targets for S-nitrosylation (7).
Several T lymphocyte functions are known to be sensitive to the GSH status (8, 9), and GSH depletion has been demonstrated in AIDS patients, suggesting the replenishment of GSH levels as a possibly useful pharmacological strategy. Increased protein glutathionylation likely plays a role in GSH depletion observed in T lymphocytes from AIDS patients (10).
35S labeling of cells has been used by Johnston and associates (11) in their earlier studies of glutathionylated protein metabolism in phagocytes. More recently, this technique was applied to study tumor necrosis factor-induced glutathionylation in HeLa cells and allowed the identification of annexin II and thioredoxin peroxidase II as proteins subject to glutathionylation (12).
We have used an approach that combines 35S labeling of intracellular glutathione followed by proteome analysis using nonreducing, two-dimensional electrophoresis and MS fingerprinting to identify specific glutathionylated proteins in human T cell blasts exposed to oxidative stress (diamide or hydrogen peroxide). This model was previously reported to cause extensive S-thiolation of cellular proteins (36).
Materials and Methods
T Cell Blasts.
Human peripheral blood mononuclear cells (PBMC) were isolated by using standard Ficoll/Hypaque gradients from buffy coats of healthy donors kindly provided by the Blood Center of the Hospital of Magenta, Magenta, Italy. PBMC were cultured at 1 × 106/ml for 3 days in RPMI 1640 with 10% FCS and 2 μg/ml phytohemagglutinin (Sigma), then washed and cultured for 3 days with 50 units/ml human recombinant IL-2 (Chiron).
Cell Labeling and Exposure to Oxidants.
Cells were resuspended at 3.3 × 106/ml and maintained throughout the experiment in Hanks' balanced salt solution (HBSS) containing 50 μg/ml cycloheximide (CHX). After a 20-min preincubation in HBSS/CHX, the cells were incubated with 8 μCi/ml of l-[35S] cysteine (specific activity, 1,000 Ci/mmol; Amersham Pharmacia). After 30 min the cells were treated for 5 min with 1 mM diamide (Sigma) or H2O2 (usually 1 mM, unless otherwise indicated). The cells were then collected by centrifugation and either processed for electrophoretic analysis as described below, or the proteins were precipitated with trichloroacetic acid (TCA) at a final concentration of 5% and filtered on GF/B glass fiber filters. After three washes with 5% TCA to remove all nonprotein-bound 35S, the amount of TCA-precipitable radioactivity was counted to assess the extent of S-thiolation induced by the oxidants.
In some experiments, the cells were treated with 10 mM buthionine sulfoximine for 30 min before and during incubation with radioactive cysteine, to inhibit the synthesis of radiolabeled GSH.
Protein Electrophoresis.
The cell pellet (2 × 107 cells/sample) was resuspended in Hanks' balanced salt solution/cycloheximide containing 50 mM N-ethylmaleimide, centrifuged, extracted with chloroform/methanol, resuspended in 8 M urea + 0.5% carrier ampholytes, and run on immobilized pH gradients (IPG) (13) covering exponentially the pH range 4 to 10 (14). IPG strips were embedded on 12% polyacrylamide gel slabs for the second-dimensional run in the discontinuous buffer system of Laemmli (15). Electroporesis was carried out under nonreducing conditions. The slabs were stained with Coomassie blue, and radioactivity was analyzed with a PhosphorImager after drying. In one experiment the protein extract of diamide-treated cells was added with 50 mM DTT and run in SDS/PAGE to confirm that no aspecific incorporation of 35S could be detected under these experimental conditions.
Protein Identification.
The 35S-labeled and Coomassie-stained protein spots were excised and washed twice during 10 min in a mixture of acetonitrile/50 mM NH4HCO3 in water (1:1 by volume). Then the gel pieces were dried in vacuo and 1 μg of trypsin in 50–100 μl of 50 mM NH4HCO3 solution was added. The digestion was allowed to proceed overnight at 37°C and was terminated by the addition of 3 μl formic acid.
The supernatant, containing the generated tryptic peptides, was transferred into another Eppendorf tube and mixed with 3 μl of a suspension of POROS 50-R2 beads in methanol (1 mg/ml). The beads preferentially adsorb the peptides, leaving buffer salts in the supernatant, which was discarded after centrifugation (16). These beads were again suspended in water, and an aliquot was placed onto the matrix-assisted laser desorption ionization-target disk and allowed to dry in the air. Peptides were desorbed on target with a solution of 4% α-cyanocinnamic acid and 1% 2,5-dihydroxybenzoic acid in 0.1% trifluoroacetic acid in water/acetonitrile (1:1 by volume).
Peptide mass fingerprints were generated with a Bruker II Reflex matrix-assisted laser desorption ionization-time of flight mass spectrometer. Proteins were identified with the mascot (Matrix Science, London) searching algorithms by using the nonredundant database (17). Further confirmation of protein identity was obtained by postsource-decay analysis of selected peptide ions and analysis with the mascot searching algorithm. Details are published elsewhere (16, 18). Because of blockage of remaining reactive sulfhydril groups by N-ethylmaleimide, we often noticed the formation of an important, and probably quantitative, amount of the N-ethylmaleimide adduct (plus 143 atomic mass units) in addition to its open hydrolyzed form (plus 125 atomic mass units). Because these groups are easily ionized, the corresponding peptides ions often appear as the dominant ions.
Enzyme Assays.
As a source of human enolase, a cytosolic preparation obtained from human PBMC was prepared by lysing PBMC (107 cells/0.1 ml) in 1% Triton X-100, 50 mM EDTA. Enolase activity was measured spectrophotometrically as described (19). Recombinant human peptidylprolyl isomerase (cyclophilin A) was a kind gift from Barbara Sherry (Picower Institute for Medical Research, Manhasset, NY). Its peptidyl-prolyl cis-trans isomerase activity was measured as published (20). Human recombinant 6-phosphogluconolactonase was prepared, and its activity was assayed as described (21).
Results
To provide a global quantitative measure of protein glutathionylation induced by exposure of T cell blasts to diamide or H2O2, we measured the amount of trichloroacetic acid-precipitable radioactivity. Five-minute exposure to 1 mM diamide resulted in a marked incorporation of radioactivity (control, 4,290 ± 1,550 cpm vs. diamide, 66,500 ± 25,000 cpm; mean ± SD of three experiments). Most (95%) of this radioactivity was attributable to S-thiolation as it was removed by DTT treatment (data not shown) and was represented by protein-bound GSH as it was reduced by 81 ± 1% if buthionine sulfoximine, an inhibitor of GSH synthesis, was added 30 min before the labeling step. A 15-min treatment with H2O2 induced S-thiolation, but to a lower extent than diamide (maximal labeling was 10,690 ± 1,360 cpm vs. 66,500 ± 25,000 cpm), with a plateau in the range of 100 μM to 10 mM H2O2 concentration. Under our experimental conditions we observed no cytotoxicity of cycloheximide, diamide, or H2O2, as judged by trypan blue exclusion (cell viability was always >90%).
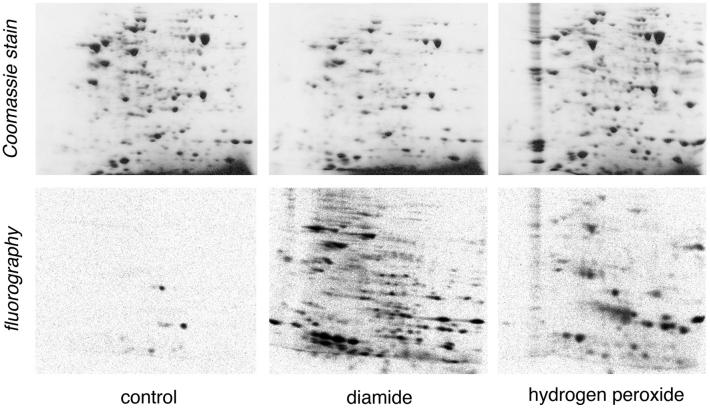
Fig. 1 shows the two-dimensional electrophoresis pattern of protein glutathionylation by diamide and H2O2 in one experiment, representative of three independent assays performed on T cell blasts obtained from different donors. Coomassie blue-stained maps are displayed in comparison with fluorography. It can be seen that a number of proteins were already labeled under basal conditions. It should be noted that this was not caused by residual protein synthesis as no radioactive spots were visible when we ran DTT-treated samples (not shown). However, in the three independent experiments, the spots labeled under basal conditions were not always the same, i.e., different proteins were found thiolated under resting conditions in different donors. As shown in Fig. 1, diamide, as expected, caused the labeling of a large number of proteins, whereas fewer proteins were labeled with H2O2. In the three experiments, the percentage of labeled proteins over Coomassie stain positive spots was 5 ± 2 in controls; 55 ± 20 in diamide-treated, and 17 ± 11 in H2O2-treated samples (mean ± SD of three experiments). Some labeled proteins, including one heavily labeled spot in control cells, were not visible in Coomassie staining and could never be identified by MS.
Figure 1.
Protein stain and glutathionylation pattern after two-dimensional electrophoresis of extracts from T cell blasts under basal conditions (control) or oxidatively stressed with 1 mM diamide or 1 mM H2O2. 35S labeling of the intracellular GSH pool and electrophoresis were performed as described in Materials and Methods. Proteins were separated based on pl (4-10 nonlinear gradient, left to right) and molecular weight (12% SDS/PAGE, top to bottom). Coomassie blue-stained maps are displayed (Upper) in comparison with fluorography (Lower). The experiment shown is representative of three and corresponds to experiment 3 as reported in Table 1.
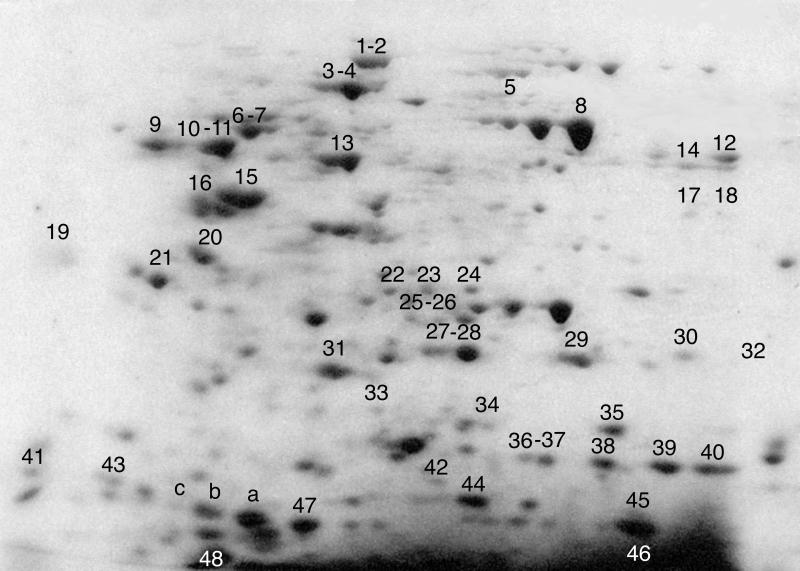
Table 1 presents an overview of all labeled proteins that could be identified by MS in one or more of the three experiments (Fig. 1 refers to experiment 3), and a semiquantitative evaluation of their labeling in the three experimental conditions (C, control; D, diamide; H, H2O2). Fig. 2 identifies the spots corresponding to the identified proteins in experiment 3 (diamide-treated cells).
Table 1.
Summary of glutathionylated proteins identified
| Protein | NCBI number | Mol mass, kDa | Spot no. (Fig. 2) | Exp. where identified by sequencing | Labeling (summary)
|
||
|---|---|---|---|---|---|---|---|
| C | D | H | |||||
| Stress proteins | |||||||
| HSP70-9B (mortalin-2) | 4758570 | 70 | 1, 2 | 3 | + | + | |
| Stress-induced phosphoprotein 1 | 400042 | 63 | 1 | + | |||
| HSP60 | 4504521 | 60 | 3, 4 | 3 | +/− | ||
| Enzymes | |||||||
| Inosine 5′-monophosphate dehydrogenase 2 | 124419 | 55 | 5 | 3 | +/− | ||
| Protein disulfide isomerase* | 1085373 | 48 | 2 | + | |||
| Enolase 1, alpha | 4503571 | 47 | 8 | 1, 2, 3 | ++/− | † | |
| (C term of) phosphoglycerate kinase | 6456828 | 12, 25 | 2‡, 3 | + | |||
| Aldolase | 229674 | 39 | 14, 17, 18 | 2§, 3 | + | ||
| 6-Phosphogluconolactonase | 6912586 | 27 | 22, 23 | 1, 3 | + | ||
| Phosphorylase kinase delta | 13544110 | 16 | 41 | 3 | + | ||
| Triosephosphate isomerase | 136060 | 26 | 1¶, 2 | + | |||
| Adenylate kinase 2 | 7434595 | 26 | 1 | ||||
| Peroxiredoxin 1 | 4505591 | 22 | 30 | 3 | + | ||
| dUTP pyrophosphatase | 1071889 | 15 | 33 | 3 | + | ||
| Nudix-type motif 6 | 11095443 | 17 | 35 | 3‖ | ++ | + | |
| Peptidylprolyl isomerase (cyclophilin A) | 10863927 | 18 | 40 | 1.3 | − | − | |
| Peptidylprolyl isomerase (cyclophilin A) | 10863927 | 18 | 36–39 | 1, 3 | + | + | |
| Cytochrome c oxidase | 4758038 | 16 | 48 | 3 | + | ||
| Ubiquitin-conjugating enzyme E2N | 4507793 | 17 | 42 | 3 | + | +/− | +/− |
| Cytoskeletal proteins | |||||||
| Vimentin | 13629179 | 53 | 6, 7 | 1, 3 | + | ||
| Vimentin (fragment?) | 13629179 | 53 | 10, 11 | 3 | + | ||
| Actin | 13278939 | 41 | 13 | 1**, 2‡‡, 3 | ++ | ++ | |
| Tropomyosin | 37424 | 29 | 19, 20 | 1, 3 | + | ||
| Transgelin, SM22 homolog calponin-like | 4507357 | 22 | 29 | 1, 2, 3 | ++ | + | |
| Cofilin | 5031635 | 18 | 34 | 1††, 3 | ++ | + | |
| Myosin | 189017 | 11 | 43 | 3 | + | ||
| Profilin | 4826898 | 15 | 45 | 3 | + | ||
| Profilin | 4826898 | 15 | 46 | 1, 2§§, 3 | + | ||
| Other | |||||||
| T complex protein 1 | 6094436 | 58 | 1¶¶ | + | |||
| Lymphocyte-specific protein 1 | 2135579 | 36 | 9 | 3 | + | ||
| Nucleophosmin (dimer?) | 13277532 | 23 | 15 | 1, 2, 3 | + | ||
| Nucleophosmin (fragment?) | 13277532 | 23 | 32 | 3 | + | ||
| Hepatoma-derived growth factor | 4758516 | 27 | 16 | 3 | + | ||
| Translation elongation factor | 4503477 | 25 | 21 | 3 | + | ||
| Endoplasmic reticulum protein | 5803013 | 29 | 23, 24 | 3 | + | ||
| SFR1 splicing factor | 730773 | 28 | 1 | + | |||
| Ash protein | 28876 | 18 | 26 | 3 | + | ||
| RNA-binding protein regulatory subunit | 6005749 | 20 | 27, 28 | 1, 3 | ++ | + | |
| My032 protein | 12002006 | 15 | 31 | 3 | ++ | ||
| Fatty acid binding protein (psoriasis assoc.) | 4557581 | 15 | 44 | 1, 2, 3 | +? | + | |
| Beta galactoside soluble lectin (galectin) | 227920 | 14 | 47∥∥ | 2, 3 | ++ | ||
| 40S ribosomal protein S12 | 133742 | 14 | 1 | +? | + | ||
NCBI, National Center for Biotechnology Information.
Weak identification.
Enolase labelling was different in the three experiments (very high in the first, null in the third).
Only in the place of spot 25 in experiment 3.
Also two low molecular weight fragments.
In the place of phosphogluconolactonase and endoplasmic reticulum protein in experiment 3 (spot 24).
In the place of cofilin in experiment 1.
One spot in the same position as experiment 3, one additional faint spot, more acidic, same molecular mass.
In a different position than experiment 3 (smaller, more basic), probably a fragment.
In a different position from experiment 3 (more basic, same molecular mass).
In a different position from experiment 3 (smaller and more acidic).
In the place of inosine 5′-monophosphate dehydrogenase 2 in experiment 3 (spot 5).
Galectin is also contained in spots a, b, and c.
Figure 2.
Two-dimensional protein map of T cell blasts. The position of the spots corresponding to the proteins identified by MS fingerprinting in experiment 3, and listed in Table 1, is marked on the map of the diamide-treated sample. Sequential numbers are either above the relevant spot (black) or below (white).
A total of 38 proteins have been identified, and almost half of them, 16, have been confirmed in at least two independent experiments. The fact that in some cases we have found the protein only once depends on the inter-individual differences on the protein pattern or on differences in the labeling pattern.
Because many of the enzymes identified as glutathionylation targets could contain cysteine residues in their active site or next to it, we investigated the effect of glutathionylation on the activity of some of them. To this purpose, recombinant enzymes, when available (peptidylprolyl isomerase and 6-phosphogluconolactonase) or a crude PBMC cytosol (as a source of enolase) were incubated for 90 min with 5 mM GSSG, or with 7.5 mM GSSG in the case of 6-phosphogluconolactonase. The effect of GSSG treatment on the activity of these enzymes is shown in Table 2. Whereas 6-phosphogluconolactonase and enolase were inhibited by glutathionylation, peptidylprolyl isomerase activity of cyclophilin was unaffected although glutathionylation of recombinant cyclophilin by GSSG was confirmed by matrix-assisted laser desorption ionization-time of flight MS (not shown).
Table 2.
Effect of GSSG on enzyme activity
| Enzyme | Without GSSG | With GSSG |
|---|---|---|
| Enolase 1, alpha* | 0.14 ± 0.02 | 0.08 ± 0.01 |
| Cyclophilin† | 0.32 ± 0.01 | 0.31 ± 0.01 |
| 6-Phosphogluconolactonase‡ | 3300 ± 100 | 1100 ± 100 |
PBMC extract (see Materials and Methods) was incubated 90 min with 5 mM GSSG, then assayed for enolase activity. Activity is expressed as μmol NADH oxidized/min per ml.
Cyclophilin (2 μM) was incubated 90 min with 5 mM GSSG, then assayed for its activity at 10°C to minimize spontaneous isomerization of the substrate. Activity is expressed as the rate of cis-trans isomerization (ΔA390/min).
Enzyme (150 μM) was incubated 90 min with 7.5 mM GSSG, then assayed for activity at 25°C. Activity is expressed as μmol/min per mg of protein.
Discussion
In this article we show that many proteins can undergo glutathionylation under oxidative stress induced by either diamide or H2O2.
A major group of glutathionylated proteins are enzymes involved in various pathways of carbohydrate/energy metabolism. This finding is in agreement with, and provides a molecular mechanism for, previous reports by Gilbert (22) who showed that the GSH/GSSG ratio regulates several enzymes of glycolysis/gluconeogenesis. While, to our knowledge, inactivation of enolase and 6-phosphogluconolactonase by GSSG had never been reported, other enzymes identified in the present study were already known to be regulated by GSSG, including aldolase (23), ubiquitin-conjugating enzyme (24, 25), triosephosphate isomerase (26), and pyrophosphatase (27). It is important to note that inactivation by GSSG does not necessarily imply glutathionylation. In fact, GSSG, by the same thiol-disulfide exchange mechanism, can also lead to formation of intrachain disulfides in the target protein, as it was elegantly shown for the OxyR transcription factor in E. coli (28). However, our study, by showing that these enzyme can incorporate 35S under oxidative stress in our experimental model, strongly suggests that the effect of GSSG reported previously is indeed caused by formation of glutathionylated proteins.
Some of the proteins found to be glutathionylated belong to the class of cytoskeletal proteins that are particularly abundant in cells. The supramolecular organization of these proteins depends on the presence of exposed -SH residues (29); the modification of these groups by glutathionylation could be relevant in their function, either by protecting them against irreversible oxidation (30) or inhibiting polymerization (31).
Other proteins identified here might have important functions. Nucleophosmin is involved in the assembly of ribosomal proteins into ribosomes and has several roles in regulating cell function and signaling (32). Cyclophilin is a chaperonin and a peptidyl prolyl isomerase whereas ubiquitin-conjugating enzyme is involved in the proteasomal degradation of proteins. Other proteins with chaperone-like activity are the heat shock proteins HSP60 and HSP70.
The consequences of glutathionylation obviously may be different for the various proteins. In the case of enzymes, we found that some of them are inhibited (enolase, 6-phosphogluconolactase) whereas for others (cyclophilin) the activity does not change with glutathionylation. In the literature, most studies report an inhibition of enzyme activity by glutathionylation but this obviously depends on the position of the cysteine undergoing modification. For instance, HIV-1 protease can be inhibited or stabilized by glutathionylation depending on which cysteine is involved (33), whereas matrix metalloproteinases are activated upon glutathionylation in the autoinhibitory domain (34).
Another important issue for which the present article can provide useful information is whether there are proteins “constitutively” glutathionylated (i.e., in the absence of oxidative stress). In our study, at least three proteins appear as a row of several spots, with different pI and the same molecular weight, which likely result from posttranslational modifications. In fact, addition of a glutamic acid residue (present in glutathione) is likely to shift toward a more acidic pI in the isoelectric focusing dimension, depending on the buffering capacity of the protein and the site of glutathionylation. In particular, cyclophilin A, containing four cysteinyl residues, has been identified in five spots, the most basic one being never labeled. Moreover the ratio between the amount of protein in the different isoforms is increased to the left in H2O2- and diamide-treated samples. This is particularly apparent in the spots marked as 38 and 39 in Fig. 2, which are almost equal in H2O2 and diamide, whereas 38 is much less abundant than 39 in control. These findings are consistent with the hypothesis that the different isoforms correspond to the progressive glutathionylation of all of the sites (from none - right, to 4 - left). Because multiple spots are present also in controls, we suggest that an extensive glutathionylation occurs even under basal conditions. A similar pattern was observed with enolase (containing six potential glutathionylation sites), but with one difference. Indeed, labeling after diamide treatment was very high in experiment 1 and zero in experiment 3, suggesting that maximal glutathionylation was already present under basal conditions in the last sample.
A different pattern can be observed for galectin, which is present as a single spot in control (47 in Fig. 2). After diamide treatment more than half of the protein disappears from this spot and appears in three more acidic spots (the last barely detectable after Coomassie blue stain; spots a, b, and c in Fig. 2). All of these spots are strongly labeled, with increasing specific activity with decreasing pI (i.e., in the presumably more extensively glutathionylated isoforms).
Taken together, these observations suggest that both basal and oxidation-induced glutathionylation occur in a significant portion of total protein molecules, supporting the hypothesis that this modification can significantly affect protein function.
In the present study, each experiment was performed by using cells obtained from a different blood donor, and it is not surprising that the redox status of the cells varies among different subjects. The fact that proteins present in large amounts, such as cyclophilin and enolase, are glutathionylated under resting conditions suggests that these proteins can also be important as glutathione stores. The hypothesis that some proteins are glutathionylated under basal conditions is in agreement with earlier reports showing that, in normal liver, 20–30 nmol of glutathione/g of liver are present as mixed disulfides with proteins (1).
It is interesting to note that H2O2 did not induce a lower labeling of the same proteins that were glutathionylated by diamide treatment but rather produced a specific labeling pattern. For instance in cyclophilin, which is already basally glutathionylated, labeling is brought about almost to the same extent by H2O2 and diamide, whereas for galectin (which is not basally glutathionylated) labeling with H2O2 is lower than with diamide, suggesting a higher reactivity of the former. It is likely that the susceptibility of different proteins to glutathionylation depends on the oxidoreduction potential of the cysteines involved, as well as on the stability of the mixed disulfide formed. For instance, it was suggested that cysteine residues flanked by basic amino acids might be particularly susceptible to glutathionylation as well as to S-nitrosylation (7, 35).
Further studies should help determine the different susceptibility of the various proteins to different degrees of oxidative stress as well as their susceptibility to being substrates for glutaredoxin and thioredoxin. The technique used here, combining proteomics and redox modification, can help identifying proteins specifically modified under pathological conditions.
Acknowledgments
We thank Dr. Barbara Sherry for the kind gift of recombinant cyclophilin. This research was supported in part by a grant (to P.G.) from the Zambon Group, Bresso, Milan, Italy. The proteome research in Ghent was supported by the Fund for Scientific Research-Flanders and by the Concerted Research Actions of the Flemish Community.
Abbreviations
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- PBMC
peripheral blood mononuclear cells
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Brigelius R, Muckel C, Akerboom T P, Sies H. Biochem Pharmacol. 1983;32:2529–2534. doi: 10.1016/0006-2952(83)90014-x. [DOI] [PubMed] [Google Scholar]
- 2.Hwang C, Sinskey A J, Lodish H F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 3.Padgett C M, Whorton A R. Am J Physiol. 1995;269:C739–C749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- 4.Cotgreave I A, Gerdes R G. Biochem Biophys Res Commun. 1998;242:1–9. doi: 10.1006/bbrc.1997.7812. [DOI] [PubMed] [Google Scholar]
- 5.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. J Biol Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura A, Yasuda K, Adachi H, Sakurai Y, Ishii N, Goto S. Biochem Biophys Res Commun. 1999;264:580–583. doi: 10.1006/bbrc.1999.1549. [DOI] [PubMed] [Google Scholar]
- 7.Jaffrey S R, Erdjument-Bromage H, Ferris C D, Tempst P, Snyder S H. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 8.Staal F J, Anderson M T, Staal G E, Herzenberg L A, Gitler C. Proc Natl Acad Sci USA. 1994;91:3619–3622. doi: 10.1073/pnas.91.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson J D, Herzenberg L A, Vasquez K, Waltenbaugh C. Proc Natl Acad Sci USA. 1998;95:3071–3076. doi: 10.1073/pnas.95.6.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Ven A J, Blom H J, Peters W, Jacobs L E, Verver T J, Koopmans P P, Demacker P, van der Meer J W. Eur J Clin Invest. 1998;28:187–193. doi: 10.1046/j.1365-2362.1998.00267.x. [DOI] [PubMed] [Google Scholar]
- 11.Rokutan K, Thomas J A, Johnston R B., Jr J Immunol. 1991;147:260–264. [PubMed] [Google Scholar]
- 12.Sullivan D M, Wehr N B, Fergusson M M, Levine R L, Finkel T. Biochemistry. 2000;39:11121–11128. doi: 10.1021/bi0007674. [DOI] [PubMed] [Google Scholar]
- 13.Gianazza E. In: 2-D Proteome Analysis Protocols. Link A J, editor. Totowa, NJ: Humana; 1998. pp. 175–188. [Google Scholar]
- 14.Gianazza E, Giacon P, Sahlin B, Righetti P G. Electrophoresis. 1985;6:53–56. [Google Scholar]
- 15.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Gevaert K, Demol H, Puype M, Broekaert D, De Boeck S, Houthaeve T, Vandekerckhove J. Electrophoresis. 1997;18:2950–2960. doi: 10.1002/elps.1150181537. [DOI] [PubMed] [Google Scholar]
- 17.Perkins D N, Pappin D J, Creasy D M, Cottrell J S. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Gevaert K, Vandekerckhove J. Electrophoresis. 2000;21:1145–1154. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1145::AID-ELPS1145>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 19.Bergmeyer H U, Grassl M, Walter H-A. In: Methods of Enzymatic Analysis. Bergmeyer H U, editor. Vol. 2. Weinheim, Germany: Verlag Chemie; 1983. pp. 182–183. [Google Scholar]
- 20.Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid F X. Nature (London) 1989;337:476–478. doi: 10.1038/337476a0. [DOI] [PubMed] [Google Scholar]
- 21.Miclet E, Stoven V, Michels P A, Opperdoes F R, Lallemand J-Y, Duffieux F. J Biol Chem. 2001;276:34840–34846. doi: 10.1074/jbc.M105174200. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert H F. Methods Enzymol. 1984;107:330–351. doi: 10.1016/0076-6879(84)07022-1. [DOI] [PubMed] [Google Scholar]
- 23.Bond J S, Offermann M K. Acta Biol Med Ger. 1981;40:1365–1374. [PubMed] [Google Scholar]
- 24.Jahngen-Hodge J, Obin M S, Gong X, Shang F, Nowell T R, Jr, Gong J, Abasi H, Blumberg J, Taylor A. J Biol Chem. 1997;272:28218–28226. doi: 10.1074/jbc.272.45.28218. [DOI] [PubMed] [Google Scholar]
- 25.Shang F, Gong X, Taylor A. J Biol Chem. 1997;272:23086–23093. doi: 10.1074/jbc.272.37.23086. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Yuksel K U, Gracy R W. Arch Biochem Biophys. 1995;317:112–120. doi: 10.1006/abbi.1995.1142. [DOI] [PubMed] [Google Scholar]
- 27.Lahti R, Suonpaa M. J Gen Microbiol. 1982;128:1023–1026. doi: 10.1099/00221287-128-5-1023. [DOI] [PubMed] [Google Scholar]
- 28.Aslund F, Beckwith J. Cell. 1999;96:751–753. doi: 10.1016/s0092-8674(00)80584-x. [DOI] [PubMed] [Google Scholar]
- 29.Stournaras C. Anticancer Res. 1990;10:1651–1659. [PubMed] [Google Scholar]
- 30.Rokutan K, Johnston R B, Jr, Kawai K. Am J Physiol. 1994;266:G247–G254. doi: 10.1152/ajpgi.1994.266.2.G247. [DOI] [PubMed] [Google Scholar]
- 31.Dalle-Donne I, Milzani A, Giustarini D, Di Simplicio P, Colombo R, Rossi R. J Muscle Res Cell Motil. 2001;21:171–181. doi: 10.1023/a:1005671319604. [DOI] [PubMed] [Google Scholar]
- 32.Chan W Y, Liu Q R, Borjigin J, Busch H, Rennert O M, Tease L A, Chan P K. Biochemistry. 1989;28:1033–1039. doi: 10.1021/bi00429a017. [DOI] [PubMed] [Google Scholar]
- 33.Davis D A, Dorsey K, Wingfield P T, Stahl S J, Kaufman J, Fales H M, Levine R L. Biochemistry. 1996;35:2482–2488. doi: 10.1021/bi951525k. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto T, Akaike T, Sawa T, Miyamoto Y, van Der Vliet A, Maeda H. J Biol Chem. 2001;276:29596–29602. doi: 10.1074/jbc.M102417200. [DOI] [PubMed] [Google Scholar]
- 35.Klatt P, Molina E P, De Lacoba M G, Padilla C A, Martinez-Galesteo E, Barcena J A, Lamas S. FASEB J. 1999;13:1481–1490. doi: 10.1096/fasebj.13.12.1481. [DOI] [PubMed] [Google Scholar]
- 36.Ghezzi P, Romines B, Fratelli M, Eberini I, Gianazza E, Casagrande S, Laragione T, Mengozzi M, Herzenberg L A, Herzenberg L A. Mol Immunol. 2002;38:773–780. doi: 10.1016/s0161-5890(01)00114-6. [DOI] [PubMed] [Google Scholar]