Abstract
The p53-related protein p73 has many functions similar to that of p53 including the ability to induce cell-cycle arrest and apoptosis. Both p53 and p73 function as transcription factors, and p73 activates expression of many genes that also are regulated by p53. Despite their similarities, it is evident that p53 and p73 are not interchangeable functionally, with p73 playing a role in normal growth and development that is not shared by p53. In this paper we describe the ability of p73β but not p53 to activate expression of the cyclin-dependent kinase inhibitor p57KIP and KvLQT1, two genes that are coregulated in an imprinted region of the genome. Our results suggest that p73 may regulate expression of genes through mechanisms that are not shared by p53, potentially explaining the different contributions of p53 and p73 to normal development.
The p53 tumor suppressor belongs to a gene family including p63 and p73, which encodes protein products with both structural and functional similarities (1). Although p53 is expressed in most tissues and plays a clear role in tumor suppression, expression of p63 and p73 seems limited to certain tissues, and their contribution to tumor development is much less well established. Neither p63 nor p73 are mutated commonly in human cancers, and mice lacking these genes are not obviously tumor prone (2–4). However, there is some evidence that inhibition of p73 activity can contribute to cancer development through deletion or methylation of the gene or interaction of the p73 protein with dominant negative forms of mutant p53 (5–10). Indeed, several tumor-associated stress signals such as DNA damage and oncogene activation that induce p53 have been shown also to activate p73 (11–16). Furthermore, unlike p53, numerous splice variants of p73 have been detected, including some isoforms that lack the N-terminal transcriptional activation domain of the protein, and thus may function to interfere with the activity of the full-length proteins (17, 18). Expression of these forms in some cancers may contribute to tumor development (19).
p73 has been shown to function as a sequence-specific transcription factor, activating expression of genes containing p53-binding sites in their promoters (20, 21). Numerous p53-inducible genes have been described, and most of them are thought to play a role in mediating the downstream effects of p53 expression. Because p73 can induce many of the genes that are activated by p53, some of the downstream responses to p73 expression such as cell-cycle arrest and apoptosis are thought to reflect this similarity in function with p53. However, despite the similarities between p53 and p73 activity, it has become evident that they are not equivalent functionally (22, 23). In comparison with p53 null mice, which generally are developmentally normal but highly tumor-susceptible, p73 null mice show several developmental defects including hydrocephalus, hippocampal dysgenesis, and inflammatory and neuronal defects (2). Although this difference may reflect a distinct expression pattern of p53 and p73 during development, it is possible also that the p73 proteins exhibit functions that are not shared by p53. Several studies have shown a quantitative difference in the ability of various p73 isoforms to induce expression of genes originally identified as p53-responsive (24–26). However, there have been few reports of activities exhibited by p73 but not shared by p53. We compared the induction of gene expression in a cell line inducibly expressing p53 and several isoforms of p73 and identified the cyclin-dependent kinase inhibitor p57KIP2 (27, 28) as a transcriptional target of p73β but not p53.
Materials and Methods
Plasmids.
Plasmids expressing N-terminal hemagglutinin (HA)-tagged human p73α, β, or γ (29) and dominant negative p73 (DDp73α; ref. 14), murine p57KIP2 (28), and human p57KIP2 (30) have been described.
Cell Lines and Transfections.
U2OS, Saos-2, H1299, and Saos-2 cells inducibly expressing p53, p73α, p73β, p73γ, and E2F1 under the control of a tetracycline-inducible promoter have been described (31, 32). Saos-E2F1-DDp73 cells were generated by transfection of the Saos-E2F1 cell line with 10 μg of pCDNA3DDp73α and 2 μg of pBabePuro (33) for stable selection of transfected cells. Cells were selected by growing in medium containing 5 μg/ml puromycin for 3 weeks, and independent clonal lines were analyzed for expression of DDp73 and retention of inducible expression of E2F1 by Western blotting. Retinal pigment epithelial cells stably expressing E1A were established after infection with an E1A-expressing retrovirus. Wild-type, androgenetic (AG), and parthenogenetic (PG) mouse embryo fibroblasts (MEFs) were derived as described (34). All cell lines were maintained in DMEM supplemented with 10% FCS and grown at 37°C in an atmosphere of 10% CO2 in air. To induce protein expression, the inducible cell lines were treated with doxycycline at 2.0 μg/ml for the indicated times. MEFs were transfected with FuGene reagent (Roche, Indianapolis) using 10 μg of DNA and 20 μl of FuGene reagent per 10-cm dish. The other cells were transfected by using the calcium-phosphate coprecipitation method.
RNA Analysis.
Total RNA was prepared with TRIzol reagent (GIBCO/BRL). Ribonuclease protection assay was performed by using 10 μg of total RNA with an RPA kit (PharMingen) and human cell cycle-2 probes labeled with [32P]UTP according to manufacturer protocol. Dried gels were scanned with a Storm PhosphorImager. For Northern blot analysis, 10 μg of total RNA per lane was separated by electrophoresis and transferred to GeneScreen Plus membrane (NEN Life Sciences). Murine p57KIP2 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH, CLONTECH) cDNA labeled with [32P]dCTP by using the Megaprime DNA labeling system (Amersham Pharmacia) for hybridization.
For reverse transcription (RT)–PCR detection of gene expression, 4 μg of total RNA was reverse-transcribed by using SuperScriptII (GIBCO). p57KIP2 cDNA was amplified with primers to exon1 (CGAACCCGACGCAGAAGAG) and exon4 (AAACAAAACCGAACGCTGCTC). GAPDH cDNA was amplified by using a human control amplimer set (CLONTECH). The PCRs were: 25 cycles of 94°C denaturation for 45 sec, 58°C annealing for 45 sec, and 72°C extension for 1.15 min. KVLQT1 cDNA was amplified by using primers to exons 5 (CCTCATCTTCTCCTCGTACTTTGTG) and 10 (GCAGTGTTGGGCTCTTCCTTACAG). LIT1 cDNA was amplified by using primers Lit105 and Lit205 described previously (35). The PCRs were: 30 cycles of 94°C denaturation for 45 sec, 52°C annealing for 45 sec, and 72°C extension for 1.15 min. The PCR products were cloned and sequenced on both strands to verify their identity.
For mRNA half-life analysis, cells were treated with doxycycline where appropriate for 20 h, and then 5 μg/ml actinomycin D was added to inhibit further transcription. Cells were harvested over a time course, and p57KIP2 mRNA levels were assessed as above.
Protein Analysis.
Western blot analysis was performed as described (36). Human HA-tagged p73α, p73β, and p73γ isoforms were detected by using F-7 anti-HA antibody (Santa Cruz Biotechnology) or rabbit polyclonal anti-p73β described previously (31). DDp73 was detected with rabbit polyclonal anti-p73α described previously (37). Human p53, MDM2, E2F1, green fluorescent protein, and β-actin were detected by using DO-1, Ab-1 (Calbiochem), anti-E2F1 KH95 (PharMingen), anti-green fluorescent protein (CLONTECH), and C4 (Chemicon) monoclonal antibodies, respectively. Human p21CIP1 was detected with EA10 (Calbiochem) or 6B6 (PharMingen) monoclonal antibodies, and human p57KIP2 was detected by using C-20 polyclonal antibodies (Santa Cruz Biotechnology).
Results
p73β Induces Expression of p57KIP2 mRNA.
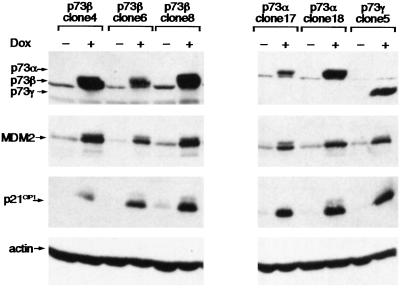
Several isoforms of p73 contain the transcriptional activation domain at the N terminus of the protein but differ in the C terminus. Although all the functional differences between these splice variants are not understood completely, it is clear that they are not equivalent in their ability to induce gene expression. For example, in several assays the p73β isoform has been found to show stronger transcriptional activity than p73α or p73γ (29), possibly reflecting an inhibitory function of the longer C-terminal region in p73α. To examine the transcriptional activity of p73 proteins we established a series of cell lines that inducibly expressed p73α, p73β, or p73γ by using the p53 null Saos-2 osteosarcoma cell line. Treatment of these cell lines with doxycycline activated expression of p73, which resulted in comparable levels of activation of two established p53 target genes, p21CIP1 and MDM2 (Fig. 1). Although some clonal variation with regard to the extent of activation could be seen, in general each of the p73 isoforms efficiently induced expression of both p21CIP1 and MDM2, which is consistent with their described ability to bind to p53-responsive promoters.
Figure 1.
Saos-2 cell lines with doxycycline-inducible expression of various p73 splice forms. Western blot analysis showing inducible expression of HA-tagged p73β, p73α, or p73γ in Saos-2 cells without (−) and after (+) treatment with 2 μg/ml doxycycline (Dox) for 40 h. p73β protein was detected by using the rabbit polyclonal antibody specific for p73β (37); p73α and p73γ were detected by using an anti-HA antibody. Induction of endogenous MDM2 and p21CIP1 is shown also. Actin expression was examined as a loading control.
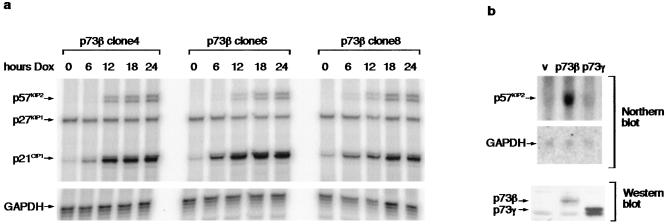
To examine the kinetics of p21CIP1 induction by p73, we carried out RNase protection. In three independent Saos-2 lines inducibly expressing p73β, p21CIP1 RNA was induced rapidly after induction of p73 by doxycycline treatment (Fig. 2a). The expression of p27KIP1, another member of this family of cyclin-dependent kinase inhibitors, remained unchanged after induction of p73β. However, we noted that expression of the third member of the cyclin-dependent kinase family, p57KIP2, also was induced in response to p73β expression in each of the three p73β-inducible clones examined. In each case, p57KIP2 expression was seen to increase between 6 and 12 h after p73β induction, somewhat later than the activation of p21CIP1, which was seen within 6 h of p73β induction. To determine whether the activation of p57KIP2 by p73β was limited to the Saos-2 cell system, we examined the effect of transfection of p73 into primary MEFs. Strong induction of mouse p57KIP2 was seen after transfection of human p73β, although a similar induction was not seen in response to p73γ despite clear expression of both proteins (Fig. 2b).
Figure 2.
Activation of p57KIP2 mRNA expression by p73β. (a) Saos-2p73 cells were untreated (0) or treated with 2 μg/ml doxycycline for the indicated times (hours Dox), and RNA was analyzed by RNase protection. Protected probes specific for p57KIP2, p27KIP1, p21CIP1, and GAPDH are shown. (b) MEFs were transiently transfected with 10 μg of vector control (v), human p73β, or human p73γ expression constructs and harvested 24 h posttransfection. (Upper) Northern blot analysis demonstrating induction of endogenous p57KIP2 is shown. GAPDH expression was examined as a loading control. (Lower) Expression of p73β and p73γ protein in the same transfected cells by Western blot using an anti-HA antibody.
p73β Induces p57KIP2 Protein Expression.
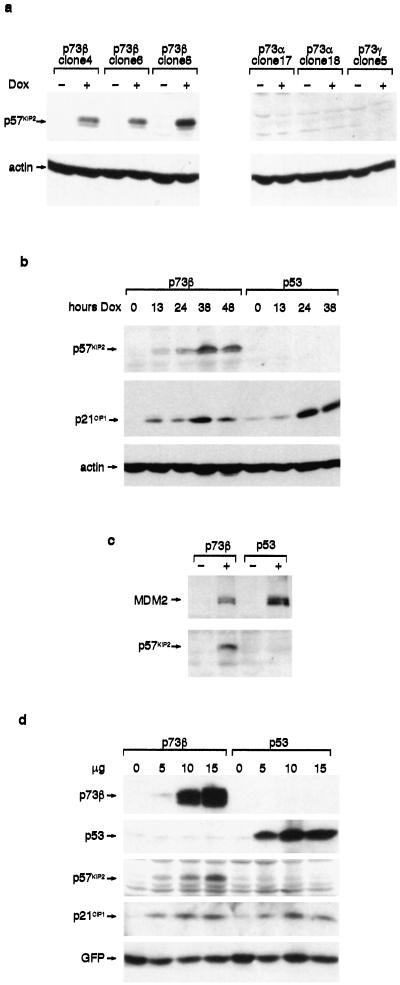
To establish that p73β expression induced p57KIP2 protein and to determine whether other p73 isoforms or p53 shared this activity, we turned back to the inducible Saos-2 cells described for Fig. 1. Induction of p73β consistently induced expression of p57KIP2 protein, but activation of p57KIP2 was not seen in response to p73α or p73γ expression (Fig. 3a) despite efficient activation of both p21CIP1 and MDM2 in each of these cell lines (Fig. 1). To examine whether this activity of p73β was shared also by p53, we used a previously published Saos-2 cell line inducibly expressing p53 (31). A time course of induction of the p73β or p53 proteins showed that they both led to the increased expression of p21CIP1, but only p73β induction led to the activation of p57KIP2 protein expression (Fig. 3b). In the same cells, induction of p53 also activated expression of MDM2 but not p57KIP2 (Fig. 3c). Transient transfection of another sarcoma cell line, U2OS, with increasing amounts of p73β also resulted in increasing amounts of both p57KIP2 and p21CIP1 protein, whereas transient transfection of p53 elevated expression of only p21CIP1 (Fig. 3d). Activation of p57KIP2 by p73β was seen also in HCT116 colon carcinoma and H1299 lung carcinoma cells (data not shown). The ability of p73β to induce p57KIP2 expression therefore was seen in human and mouse cells, tumor cell lines, and primary untransformed fibroblasts.
Figure 3.
Activation of p57KIP2 protein expression by p73β. (a) Various Saos-2-p73 cells were untreated (−) or treated (+) with doxycycline (Dox) for 40 h as described for Fig. 1, and p57KIP2 protein expression was analyzed by Western blotting as indicated. (b) Saos-2-p73 and Saos-2-p53 cells were untreated (0) or treated with 2 μg/ml doxycycline (hours Dox) for the indicated times, and p57KIP2 and p21CIP1 protein was analyzed by Western blotting. Actin expression was examined as a loading control. (c) Saos-2-p73β and Saos-2-p53 cells were untreated (−) or treated (+) with doxycycline for 40 h, and p57KIP2 and MDM2 protein were analyzed by Western blotting. (d) U2OS cells were transfected with 15.5 μg of DNA consisting of 0, 5, 10, or 15 μg of p53 or p73β expression plasmids (as indicated) supplemented with vector control DNA, and 0.5 μg of green fluorescent protein (GFP) was added as a transfection efficiency control. Protein expression was analyzed as described above; p73β was detected by using the p73β-specific polyclonal antibody, and p53 was detected by using the p53-specific monoclonal antibody DO-1.
E2F1 Induces p57KIP2 in a p73-Dependent Manner.
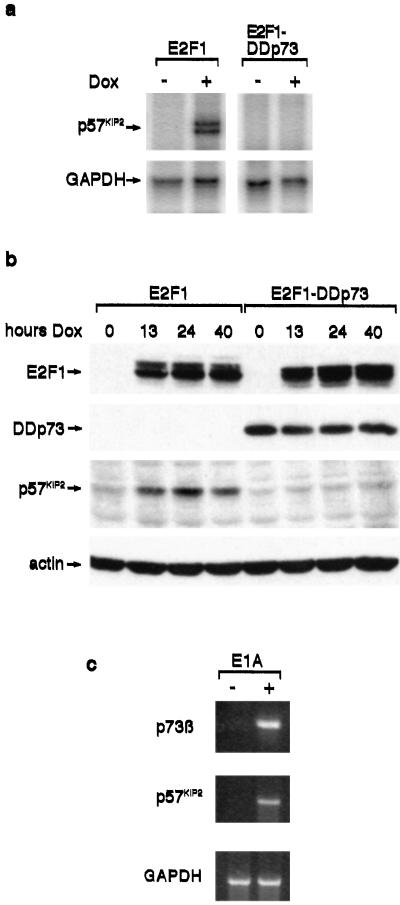
Although many previous studies have identified p53-target genes that also are transcriptionally activated in response to p73, it has been pointed out that most of these studies have depended on overexpression of exogenous p73 in cells (23). To determine whether endogenous p73β could lead to an increase in p57KIP2 expression, we used another Saos-2 cell line inducibly expressing E2F1 (38). Induction of E2F1 in this cell line had been shown previously to result in elevated expression of endogenous p73α and p73β, with evidence that the E2F1-induced apoptotic response at least in part depended on this activation of the p73 proteins (14). Consistent with the observation that p73β can activate expression of p57KIP2, we found by RNase protection that induction of E2F1, leading to activation of p73β, resulted in an increased expression of p57KIP2 (Fig. 4a). Expression of a dominant negative form of p73 previously shown to inhibit the activity of p73 proteins (DDp73; ref. 14) abolished the induction of p57KIP2 expression, supporting a role for p73β in mediating E2F1-dependent activation of p57KIP2. Similar results were obtained when protein expression in these cells was examined by Western blotting (Fig. 4b). Induction of E2F1 led to the elevation of p57KIP2 protein, which was blocked by expression of the dominant negative p73 protein in these cells. To determine whether endogenous E2F1 could have the same effect, we examined primary human retinal pigment epithelial cells expressing E1A, a viral protein that binds the pRB family and deregulates the activity of the E2F (39). Compared with mock-infected cells, E1A expressing primary human retinal pigment epithelial cells showed elevated expression of both p73β and p57KIP2 (Fig. 4c). Taken together, the results indicate that endogenous p73β expression can lead to the activation of p57KIP2 expression.
Figure 4.
E2F1 induction of p57KIP2 is p73-dependent. (a) Saos-E2F1 and Saos-E2F1-DDp73 cells were untreated (−) or treated (+) with 2 μg/ml doxycycline (Dox) for 12 h, and total RNA was analyzed by RNase protection. Protected probes specific for p57KIP2 and GAPDH are shown. (b) Saos-E2F1 and Saos-E2F1-DDp73 cells were untreated (0) or treated with doxycycline for the indicated times (hours Dox). Expression of E2F1, DDp73, and p57KIP2 proteins was analyzed by Western blotting. Actin expression was examined as a loading control. (c) Primary human retinal pigment epithelial cells without (−) or with (+) E1A were examined for p73β and p57KIP2 mRNA expression levels by RT-PCR. GAPDH expression was included as a loading control.
Regulation of KvLQT Expression by p73β.
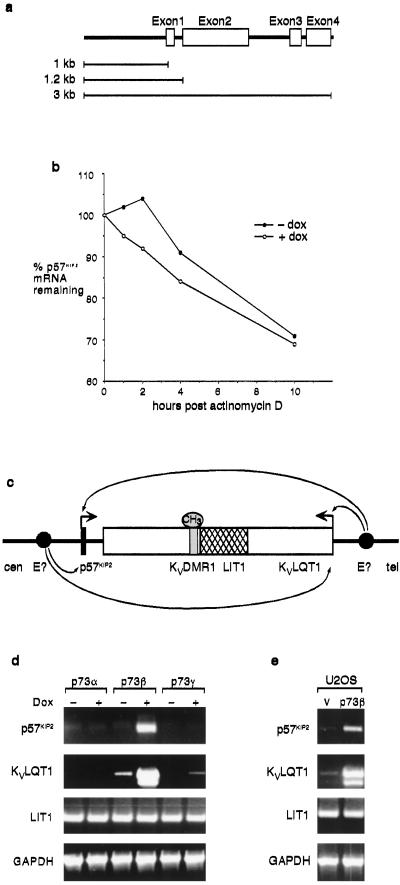
p73β is a transcription factor that can show sequence-specific DNA binding, and thus we sought to identify a p73-dependent element in the p57KIP2 promoter. Previous studies have shown that the 165-bp region upstream of the human p57KIP2 transcription initiation site contains binding sites for numerous transcription factors including SP1, NF1, OCT1, EGR1, TBP, and ETS (40). This fragment functioned as a promoter and could be repressed by EWS-FLI-1. Analysis of the sequences upstream of the p57KIP2 transcriptional start or in the introns of the p57KIP2 gene (Fig. 5a) failed to reveal any p53-binding sites arranged in tandem, as usually is seen in p53-responsive promoters. Because we had not found p57KIP2 to be obviously p53-responsive, the absence of a consensus p53-binding site was not surprising. We cloned 1 kb of upstream sequences and the sequences contained in the first intron of human p57KIP2into a luciferase reporter construct to examine possible p73 responsiveness. However, although these reporter constructs showed high constitutive expression after transfection into cells, we were unable to measure any p73β-dependent increase in activity (data not shown). To identify an enhancer element within the p57KIP2 gene, we cloned the whole 3-kb genomic region downstream of the luciferase gene driven by a simian virus 40 promoter. Again, we failed to detect any p73β-dependent increase in luciferase activity. To examine the possibility that p73β expression affected p57KIP2 mRNA stability, we examined the half-life of p57KIP2 mRNA in response to p73β expression (Fig. 5b). These experiments showed that the half-life of the p57KIP2 mRNA was not extended by expression of p73β, supporting the hypothesis that p73β expression leads to up-regulation of p57KIP2 transcription.
Figure 5.
p57KIP2 and KvLQT1 are regulated coordinately by p73β. (a) Genomic organization of the p57KIP2 gene. The sequences studied by luciferase reporter assays are indicated as 1 kb, 1.2 kb, and 3 kb. (b) Saos-2p73β cells were untreated (− dox) or treated (+ dox) with 2 μg/ml doxycycline for 20 h. Cells were harvested at the indicated time points after the addition of 5 μg/ml actinomycin D, and the remaining p57KIP2 mRNA levels were assessed. (c) Regulation of p57KIP2 and KvLQT1 by a putative shared enhancer (E?, closed circle). The drawing represents ≈500 kb of the genome located on chromosome 11p15.5. Closed, open, and hatched boxes represent the p57KIP2, KvLQT1, and LIT1 genes, respectively, small arrows show gene activation, and large arrows show the direction of transcription. Cen and tel refer to the orientation of the centromere and telomere, respectively. (d) Saos-2p73α, p73β, and p73γ cells were untreated (−) or treated (+) with 2 μg/ml doxycycline (Dox) for 12 h, and RNA was analyzed by RT-PCR. (e) U2OS cells were transfected with 10 μg of vector control (v) or p73β expression plasmid. Twenty-four hours after transfection, total RNA was analyzed by RT-PCR.
The p57KIP2 gene is localized to the Beckwith–Wiedemann syndrome (BWS) cluster on chromosome 11p15.5, and loss of p57KIP2 expression may play a causative role in BWS (41, 42). This chromosomal region contains a major imprinted gene cluster with coordinately and oppositely imprinted genes. Although the exact mechanisms by which expression of this gene cluster is regulated remain to be determined, it has been suggested that the p57KIP2 gene and an adjacent gene, KvLQT1, may be regulated coordinately from the same enhancer, which may be telomeric or centromeric to KvLQT1 (refs. 43 and 44; Fig. 5c). The KvLQT1 gene is over 400 kb long and encodes an ion channel that is mutated in syndromes associated with heart disease and deafness (45). Because we were not able to identify a region directly within the p57KIP2 promoter that was p73β-responsive, we considered the possibility that p73β regulates p57KIP2 through the enhancer shared with the KvLQT1 gene. By using RT-linked PCR, we found that similar to p57KIP2, KvLQT1 was induced after expression of p73β (Fig. 5 d and e). These results suggest that p73β may play a role in overall regulation of these genes in the BWS cluster.
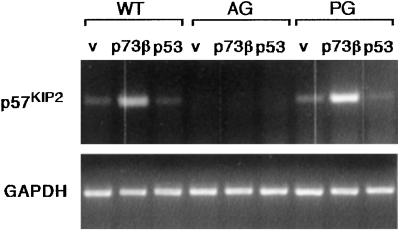
Expression of both p57KIP2 and KvLQT1 can be regulated by differential methylation of the KvDMR1 site within KvLQT1 (Fig. 5c). Methylation of this site in the normal maternal chromosome allows maternal expression of these genes. The LIT1 gene, which is localized within the KvLQT1 gene, is expressed in the reverse orientation from KvLQT1 from the paternal allele, where the KvDMR site is not methylated (46, 47). If p73β expression regulated the methylation of the KvDMR1 site, we would predict that expression of LIT1 would be repressed as p57KIP2 and KvLQT1 expression was activated. However, we found that expression of the LIT1 gene was not affected after expression of p73β in either Saos-p73β or U2OS cells (Fig. 5 d and e). Control PCRs, in which RNA was treated in the same conditions without addition of reverse transcriptase, did not yield any product, showing that the RNA is free of contamination with genomic DNA (data not shown). Furthermore, we found that p73β expression did not alter the expression of UBE3A, a maternally imprinted gene on human chromosome 15, that is linked to Angelman syndrome (ref. 48; data not shown). These results suggested that p73β expression does not regulate expression of all imprinted genes globally. To study this result more closely, we examined the effect of p73β expression in uniparental MEFs containing exclusively either the paternal (AG) or maternal (PG) genome. These cells have been shown to retain the correct expression of imprinted genes, and thus p57KIP2 is expressed only in the maternally derived PG cells (34). Expression of p73β in the AG cells did not induce any detectable p57KIP2 expression (Fig. 6), although previous studies have shown that treatment of these cells with the histone deacetylate inhibitor TSA or the DNA methyltransferase inhibitors AzaC and AzadC resulted in activation of p57KIP2 expression (34). Interestingly, expression of p73β did enhance the expression of p57KIP2 significantly in the PG cells, in which neither allele is silenced and basal levels of p57KIP2 expression are slightly higher than that detected in wild-type cells (Fig. 6; ref. 34). These results show that p73β is activating expression of the nonsilenced p57KIP2 allele.
Figure 6.
p73β cannot activate expression of p57KIP2 in uniparental AG MEFs. Retroviral infection was used to achieve efficient expression of p73β or p53 in wild-type (WT), AG, or PG MEFs. Cells were harvested 24 h postinfection, and expression of p57KIP2 mRNA was analyzed by RT-PCR. GAPDH expression was included as a loading control.
Interestingly, when studying the induction of p57KIP2 in Saos-p73β and transiently transfected U2OS cells by using RT-PCR, we were able to detect production of both full-length p57KIP2 mRNA and alternatively spliced forms depending on the primer pair used (data not shown). One of the predominant alternatively spliced forms has been described (27), and would be predicted to lead to the expression of a protein with a different C terminus because of loss of the 3′ end of exon 2 and a resulting frame shift in exon 3. The expression of this variant protein was not detected, because the antibody we used for Western blotting (Fig. 3) was raised against the C terminus of the full-length p57KIP2 protein.
Discussion
In this study we identify the cyclin-dependent kinase inhibitor p57KIP2 as a target for p73β-mediated transcriptional activation. Other isoforms of p73 (p73α and p73γ) fail to induce p57KIP2. The differential ability of various p73 isoforms to induce target gene expression has been described in previous studies (24–26, 29). We were unable to detect induction of p57KIP2 by p53, but it remains possible that in some cell types p53 also will activate expression of this gene.
Although we have been able to detect activation of p57KIP2 by p73β in both human and mouse cells and in primary cells and cancer cells lines, and in response to exogenous and endogenous p73β expression, we thus far have been unable to determine the mechanism by which p73β induces expression of p57KIP2. Our attempts to delineate a p73-responsive element in the upstream or first-intron sequences of p57KIP2 have failed, although it is possible that we have not extended our analysis sufficiently or the activity was masked by the high constitutive expression of the reporter constructs. Nevertheless, we found that the activation of expression in response to p73β was not limited to p57KIP2 but also shared by KvLQT1, another gene known to be coregulated in this imprinted region of the Beckwith–Wiedemann syndrome cluster (43, 44). These results suggest that p73β may play a role in regulating the coordinate expression of these imprinted genes, although the failure of p73β to enhance expression of other imprinted genes and the inability of p73β to activate expression of p57KIP2 in uniparental AG MEFs suggest that p73β is not regulating the methylation or acetylation status of the silenced alleles. The ability of p73β to further enhance expression of p57KIP2 in the PG cells suggests that p73β may be controlling the activity of the proposed p57KIP2/KvLQT1 enhancer, which could lie several hundred kilobases from the p57KIP2 gene itself. Regulation of p57KIP2 expression may be a direct or indirect activity of p73β and could represent functions not shared by p53.
To date, the best understood activities of p73β are those shared by p53, namely the ability to induce cell-cycle arrest and apoptosis in cultured cells. Because p57KIP2 is a cyclin-dependent kinase inhibitor (27, 28), it is possible that the induction of p57KIP2 in response to p73β plays a role in the activation of a cell-cycle arrest. However, p73β, similar to p53, also induces expression of p21CIP1, and it seems unlikely that p57KIP2 is the sole mediator of cell-cycle arrest. Our data suggest that p57KIP2 activation will reflect an activity of p73β not shared by p53, and it is of interest to note that unlike p21CIP1, p57KIP2 can play a role in development (42, 49). Developmental defects also are seen in mice deficient in the KvLQT1 gene, which exhibit gastric hyperplasia as well as complete deafness, imbalance, and disruption of the cochlear and vestibular organs (50). Although the phenotype of p57KIP2-, KvLQT1-, and p73-deficient mice are not obviously similar, it seems possible that the developmental defects seen in mice that lack p73 may reflect, in part, the loss of normal activation of p57KIP2 and KvLQT1.
Acknowledgments
We thank Gerry Melino, William Kaelin, and Takashi Tokino for the kind gift of plasmids.
Abbreviations
- HA
hemagglutinin
- AG
androgenetic
- PG
parthenogenetic
- MEF
mouse embryo fibroblast
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kaelin W G., Jr Oncogene. 1999;18:7701–7705. doi: 10.1038/sj.onc.1202955. [DOI] [PubMed] [Google Scholar]
- 2.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. Nature (London) 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 3.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson R T, Tabin C, Sharpe A, Caput D, Crum C, McKeon F. Nature (London) 1999;398:714–717. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 4.Mills A A, Zheng B, Wang X-J, Vogel H, Roop D R, Bradley A. Nature (London) 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 5.Kawano S, Miller C W, Gombart A F, Bartram C R, Matsuo Y, Asou H, Sakashita A, Said J, Tatsumi E, Koeffler H P. Blood. 1999;94:1113–1120. [PubMed] [Google Scholar]
- 6.Corn P G, Kuerbitz S J, van Noesel M M, Esteller M, Compitello N, Baylin S B, Herman J G. Cancer Res. 1999;59:3352–3356. [PubMed] [Google Scholar]
- 7.Di Como C J, Gaiddon C, Prives C. Mol Cell Biol. 1999;19:1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marin M C, Jost C A, Brooks L A, Irwin M S, O'Nions J, Tidy J A, James N, McGregor J M, Harwood C A, Yulug I G, et al. Nat Genet. 2000;25:47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 9.Strano S, Munarriz E, Rossi M, Cristofanellu B, Shaul Y, Castagnoli L, Levine A J, Sacchi A, Cesareni G, Oren M, Blandino G. J Biol Chem. 2000;275:29503–29512. doi: 10.1074/jbc.M003360200. [DOI] [PubMed] [Google Scholar]
- 10.Gaiddon C, Lokshn M, Ahn J, Zhang T, Prives C. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agami R, Blandino G, Oren M, Shaul Y. Nature (London) 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 12.Gong J G, Costanzo A, Yang H-Q, Melino G, Kaelin W G, Jr, Levrero M, Wang J Y J. Nature (London) 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Z-M, Shioya H, Ishiko T, Sun X, Gu J, Huang Y Y, Lu H, Kharbanda S, Weichselbaum R, Kufe D. Nature (London) 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 14.Irwin M, Marin M C, Phillips A C, Seelan R S, Smith D I, Liu W, Flores E R, Tsai K Y, Jacks T, Vousden K H, Kaelin W G., Jr Nature (London) 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 15.Stiewe T, Putzer B M. Nat Genet. 2000;26:464–469. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 16.Zaika A, Irwin M, Sansome C, Moll U. J Biol Chem. 2001;276:11310–11316. doi: 10.1074/jbc.M005737200. [DOI] [PubMed] [Google Scholar]
- 17.Filloppovich I, Sorokina N, Gatei M, Haupt Y, Hobson K, Moallem E, Spring K, Mould M, McGuckin M A, Lavin M F, Khanna K K. Oncogene. 2001;20:514–522. doi: 10.1038/sj.onc.1204118. [DOI] [PubMed] [Google Scholar]
- 18.Grob T J, Novak U, Maisse C, Barcaroli D, Lüthi A U, Pirnia F, Hügli B, Graber H U, De Laurenzi V, Fey M F, Melino G, Tobler A. Cell Death Diff. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- 19.Casciano I, Ponzoni M, Cunsolo C, Tonini G, Romani M. Cell Death Diff. 1999;6:391–393. doi: 10.1038/sj.cdd.4400522. [DOI] [PubMed] [Google Scholar]
- 20.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J-C, Valent A, Minty A, Chalon P, Lelias J-M, Dumont X, Ferrara P, McKeon F, Caput D. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 21.Jost C A, Marin M C, Kaelin W G., Jr Nature (London) 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 22.Levrero M, de Laurenzi V, Costanzo A, Sabatini S, Gong J, Wang J Y J, Melino G. J Cell Biol. 2000;113:1661–1670. doi: 10.1242/jcs.113.10.1661. [DOI] [PubMed] [Google Scholar]
- 23.Irwin M S, Kaelin W G., Jr Cell Growth Diff. 2001;12:337–349. [PubMed] [Google Scholar]
- 24.Zhu J, Jiang J, Zhou W, Chen X. Cancer Res. 1998;58:5061–5065. [PubMed] [Google Scholar]
- 25.Lee C-W, La Thangue N B. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Zhang L, Hwang P M, Rago C, Kinzler K, Vogelstein B. Proc Natl Acad Sci USA. 1999;96:14517–14522. doi: 10.1073/pnas.96.25.14517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuoka S, Edwards M C, Bai C, Parker S, Zhang P, Baldini A, Harper J W, Elledge S J. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 28.Lee M, Reynisdottir I, Massague J. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 29.De Laurenzi V, Constanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. J Exp Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokino T, Urano T, Furuhata T, Matsushima M, Miyatsu T, Sasaki S, Nakamura Y. Hum Genet. 1996;97:625–631. doi: 10.1007/BF02281873. [DOI] [PubMed] [Google Scholar]
- 31.Nakano K, Balint E, Ashcroft M, Vousden K H. Oncogene. 2000;19:4283–4289. doi: 10.1038/sj.onc.1203774. [DOI] [PubMed] [Google Scholar]
- 32.Phillips A C, Ernst M K, Bates S, Rice N R, Vousden K H. Mol Cell. 1999;4:771–781. doi: 10.1016/s1097-2765(00)80387-1. [DOI] [PubMed] [Google Scholar]
- 33.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Kharroubi A, Piras G, Stewart C L. J Biol Chem. 2001;276:8674–8680. doi: 10.1074/jbc.M009392200. [DOI] [PubMed] [Google Scholar]
- 35.Lee M P, DeBaun M R, Mitsuya K, Galoek H L, Brandenburg S, Oshimura M, Feinberg A P. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashcroft M, Taya Y, Vousden K H. Mol Cell Biol. 2000;20:3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balint E, Bates S, Vousden K H. Oncogene. 1999;18:3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 38.Phillips A C, Bates S, Ryan K M, Helin K, Vousden K H. Genes Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- 39.Vousden K H. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 40.Dauphinot L, De Oliveira C, Melot T, Sevenet N, Thomas V, Weissman B E, Delattre O. Oncogene. 2001;20:3258–3265. doi: 10.1038/sj.onc.1204437. [DOI] [PubMed] [Google Scholar]
- 41.Matsuoka S, Thompson J S, Edwards M C, Barletta J M, Grundy P, Kalikin L M, Harper J W, Elledge S J, Feinberg A P. Proc Natl Acad Sci USA. 1996;93:3026–3030. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Liégeois N J, Wong C, Finegold M, Hou H, Thompson J C, Silverman A, Harper J W, DePinho R A, Elledge S J. Nature (London) 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- 43.Maher E, Reik W. J Clin Invest. 2000;105:247–250. doi: 10.1172/JCI9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feinberg A P. J Clin Invest. 2000;106:739–740. doi: 10.1172/JCI10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Bowles N E, Towbin J A. Mol Med Today. 1998;4:382–388. doi: 10.1016/s1357-4310(98)01320-3. [DOI] [PubMed] [Google Scholar]
- 46.Mitsuya K, Meguro M, Lee M P, Katoh M, Schulz T C, Kugoh H, Yoshida M A, Niikawa N, Feinberg A P, Oshimura M. Hum Mol Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 47.Smilinich N J, Day C D, Fitzpatrick G V, Caldwell G M, Lossie A C, Cooper P R, Smallwood A C, Joyce J A, Schofield P N, Reik W, et al. Proc Natl Acad Sci USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholls R D, Kneppwe J L. Annu Rev Genome Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y, Frisén J, Lee M-H, Massahué J, Barbacid M. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- 50.Lee M P, Ravenel J D, Hu R-J, Lustig L R, Tomaselli G, Berger R D, Brandenburg S A, Litzi T J, Bunton T E, Limb C, et al. J Clin Invest. 2000;106:1447–1455. doi: 10.1172/JCI10897. [DOI] [PMC free article] [PubMed] [Google Scholar]