Abstract
Elastic distortion of a structural element of the actomyosin complex is fundamental to the ability of myosin to generate motile forces. An elastic element allows strain to develop within the actomyosin complex (cross-bridge) before movement. Relief of this strain then drives filament sliding, or more generally, movement of a cargo. Even with the known crystal structure of the myosin head, however, the structural element of the actomyosin complex in which elastic distortion occurs remained unclear. To assign functional relevance to various structural elements of the myosin head, e.g., to identify the elastic element within the cross-bridge, we studied mechanical properties of muscle fibers from patients with familial hypertrophic cardiomyopathy with point mutations in the head domain of the β-myosin heavy chain. We found that the Arg-719 → Trp (Arg719Trp) mutation, which is located in the converter domain of the myosin head fragment, causes an increase in force generation and fiber stiffness under isometric conditions by 48–59%. Under rigor and relaxing conditions, fiber stiffness was 45–47% higher than in control fibers. Yet, kinetics of active cross-bridge cycling were unchanged. These findings, especially the increase in fiber stiffness under rigor conditions, indicate that cross-bridges with the Arg719Trp mutation are more resistant to elastic distortion. The data presented here strongly suggest that the converter domain that forms the junction between the catalytic and the light-chain-binding domain of the myosin head is not only essential for elastic distortion of the cross-bridge, but that the main elastic distortion may even occur within the converter domain itself.
It is widely accepted that active force and movement generated by muscle fibers result from structural changes in the head domain of the myosin molecule (the cross-bridge) while it is attached to the actin filament. These changes are thought to involve a tilting of the light-chain-binding domain of the myosin head relative to its catalytic domain (1–7). As a result of such structural changes distortion of an elastic element within the actin-attached myosin head allows strain to develop before movement (8). Relief of this strain drives sliding of actin filaments past the myosin filaments, or alternatively, if filament sliding is prevented, active force is generated because of continued strain of the elastic element. Despite the central significance of this concept, however, it remained unclear which part of the actomyosin complex represents the elastic element—i.e., the element that experiences the main elastic distortion while other parts act more like rigid bodies. Neither the known crystal structures of the myosin head nor cryo-electron microscopy with reconstruction of the actomyosin complex have resolved this question. Some authors considered elastic bending of the long, light-chain-binding α-helix an obvious candidate (4, 9, 10). The actin–myosin interface (11) or the junction between the light-chain domain and the catalytic domain of the myosin head (9) as well as subfragment 2 were also considered (12) as other possibilities for the location of elastic distortion.
To identify the structural element of the myosin head domain that acts as the main elastic element we made use of missense mutations in the head domain of the β-myosin heavy chain (β-MHC), the heavy chain isoform that is expressed in cardiac and in slow skeletal muscle. Such myosin mutations were shown to be associated with familial hypertrophic cardiomyopathy (FHC) in about 30% of all kindreds with this disease (13–15). In this study we examined fibers from a soleus muscle biopsy of a patient with a substitution of arginine by tryptophane at position 719 of the myosin heavy chain (16). This mutation is located within the converter domain of the myosin head (Fig. 1) which extends from Phe-707 to Arg-774, as defined by Houdusse and coworkers on the basis of the crystal structure of scallop striated muscle myosin subfragment-1 (17). The converter domain forms a small compact subdomain that links the α-helical light-chain-binding domain to the catalytic domain (7, 17, 18). The converter and light-chain-binding domain were thought to act together as a (semirigid) lever arm (2–7, 17), but the structural element that becomes elastically distorted remained unclear. Our results show that the converter domain seems to be a key element of the actomyosin complex where elastic distortion occurs.
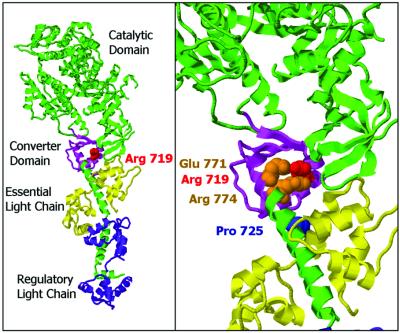
Figure 1.
Ribbon diagram of the structure of the scallop striated muscle subfragment-1 (17) showing the position of the FHC mutation studied here. (Left) Heavy chain of S1 (catalytic domain pointing to the upper left) and the long α-helix representing the lever arm are in green, light chains in magenta and yellow, the converter domain in light violet. The converter domain encompasses residues Phe-707 to Arg-774 (17). Arg-719 is displayed space-filling in red. The crystal structure shown here indicates the very close proximity of the mutated residue and the first turns of the α-helix forming the core of the light-chain-binding domain. (Right) Close-up view of the converter area highlighting two residues of the long α-helix (Glu-771 and Arg-774, space-filling in orange) that are in close contact with Arg-719, which has been described to be conserved in all heavy-chain isoforms (17). Pro-725 of the converter is shown space-filling in blue to illustrate the part of the converter that is close to the essential light chain. (Figure prepared with RasMol.)
Materials and Methods
Fiber Preparation and Solutions.
Soleus muscle biopsies were obtained under local anesthesia from a female member of a British family with the Arg719Trp mutation in the β-MHC and from four control individuals undergoing plastic surgery. Informed consent of the individuals was obtained according to approved Ethics Committee protocols of the St. George's Hospital, London and of the Medical School, Hannover. All biopsies were dissected into small muscle fiber bundles immediately after surgery and permeabilized in skinning solution with Triton-X-100 (19). For long-term storage, bundles of permeabilized fibers were equilibrated for 1 h each in skinning solution containing 0.5, 1.0, 1.5, and finally 2.0 M sucrose, rapidly frozen in liquid propane, and stored in liquid N2 (20). To isolate single fibers, after thawing the bundles were reequilibrated in each of these sucrose concentrations but in reversed order before transfer into sucrose-free skinning solution. Care was taken that during the entire dissection and equilibration procedure the muscle preparation was kept at low temperature (2–5°C). Skinning solution and rigor, activating, and relaxing solutions were as described (19) except for the addition of 5 mM 2,3-butanedione monoxime to the skinning solution and the relaxing solutions. To keep fibers fully relaxed, even at higher temperatures despite the shallow force-calcium relation, 2,3-Butanedione monoxime was added (ref. 21; T.K., unpublished results). Measurements in rigor and active contraction were performed at 170 mM ionic strength. In relaxation an ionic strength of 50 mM was chosen to favor weak cross-bridge attachment. Stiffness in rigor and relaxation was measured at 5°C, active parameters at 10°C and 20°C.
Mechanical and X-Ray Diffraction Experiments.
Single fibers were isolated and mounted into the setup as described (19, 22). The cross-sectional area of the fibers was first determined by light microscopy assuming an elliptical shape of the fibers. To obtain more precise values for force and stiffness per cross-sectional area of mutant and control fibers, the cross-sectional area of most fibers was also measured from optical cross-sections recorded with a confocal microscope integrated into our mechanical setup. To record optical cross-sections, fibers were labeled with rhodamine-phalloidin in relaxing solution after mechanical measurements were completed. To document structural integrity of fibers and homogenous packing of myofibrils in fibers of both controls and patient, longitudinal optical sections were also recorded (see Fig. 3).
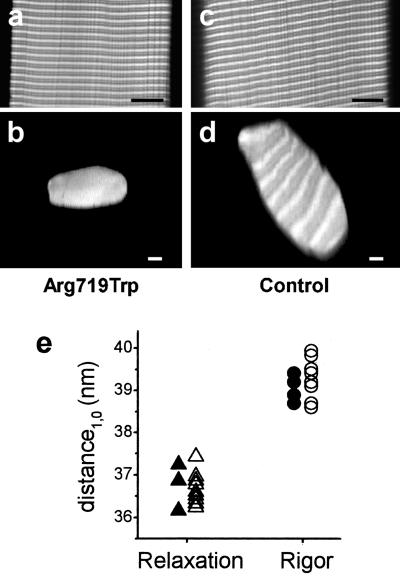
Figure 3.
Confocal images and x-ray diffraction data to ensure precise measurement of fiber cross-sections and normal content of myofibrils and myofilaments in both fibers with mutant myosin and control fibers. (a and c) Longitudinal optical sections through the core of the fibers; (b and d) optical cross-sections of the same fibers. (Left) Fiber with mutated myosin; (Right) control fiber. (Scale bars: 10 μm.) Fibers were labeled with rhodamine-phalloidin, which under the conditions used here binds to the actin filaments in the overlap region and near the Z-line. The light and dark pattern of the cross-sections arises form cutting through unlabeled and labeled regions of the sarcomere. (e) Distance of the two innermost equatorial reflections (d1,0) in x-ray diffraction patterns obtained in rigor and relaxation from fibers with mutated myosin (open symbols) and control fibers (filled symbols).
Resistance to elastic distortion (i.e., fiber stiffness) was measured with ramp-shaped stretches or releases of different speeds [10−1 to 3 × 103 (nm/half-sarcomere) s−1] with change in sarcomere length recorded by laser light diffraction (23). Stiffness was determined as chord stiffness—i.e., from the force change when the sarcomere length change had reached 2 nm/half-sarcomere. Actively contracting fibers were stabilized by a quick release/restretch protocol (24) and the rate constant of force redevelopment (kredev) was determined after the restretch to isometric fiber length (25). Unloaded shortening velocity (vmax) was determined by slack tests (26).
To determine the distance between thick and thin filaments and between crowns of myosin heads along the myosin filaments, the spacings of the 1.0 equatorial reflection (d1,0) and the 3rd meridional reflection (dM3) were determined from x-ray diffraction patterns. The patterns were recorded at the Deutsches Elektronen Synchrotron in Hamburg, Germany as described (27), except that single fiber arrays of only 8–20 fibers were used because these were sufficient to record the necessary meridional and equatorial reflections. Solutions and temperature were the same as for mechanical experiments except for the addition of 10 mM glutathione and 1,000 units/ml catalase to reduce damage by the x-ray irradiation.
Histological and Biochemical Analysis.
Part of each biopsy was subjected to routine histochemical and immune-histochemical analysis, including ATPase and NADH reactions, as well as oil-red and hematoxylin-eosin staining. All samples had normal enzymatic patterns and normal structure. The myosin heavy-chain isoforms were separated by using SDS/PAGE (28) to confirm that only fibers with the slow β-MHC isoform exclusively were included in the analysis of the functional properties. For this purpose, after analysis of functional properties and optical cross-section, each fiber was immediately transferred into SDS buffer and frozen for later electrophoresis.
Results
Force and Fiber Stiffness in Active Isometric Contraction.
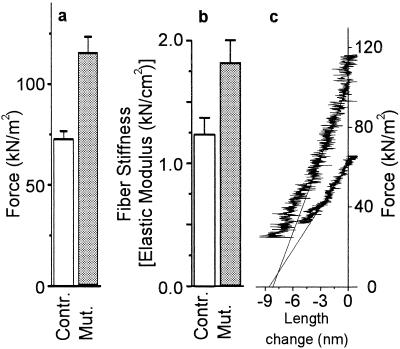
To characterize functional effects of the Arg719Trp mutation we first measured active force and fiber stiffness (resistance to elastic distortion) under isometric conditions. At full calcium activation we found an increase in active force by 59 ± 7% at 10°C (Fig. 2a) and by 49 ± 5% at 20°C compared with isometric force of slow soleus muscle fibers from healthy control individuals. Fiber stiffness under isometric conditions at 10°C was 48 ± 10% higher than with normal controls, when length changes (stretches or releases) with speeds of ≈2 × 103 (nm/half-sarcomere) s−1 were imposed (Fig. 2b). As an example, Fig. 2c shows two original records from which stiffness of activated fibers was determined. In these records active force is plotted against the change in sarcomere length during releases which were imposed during isometric steady state contraction. The solid lines were fitted to the linear part of these plots. The slopes of these lines represent apparent fiber stiffness. Note that the x axis intercept [y0 value (29)] is almost identical for these two individual plots. The average values of all x axis intercepts were 7.09 ± 0.57 nm/half-sarcomere (n = 14) and 7.66 ± 0.99 nm/half-sarcomere (n = 9) for control and fibers with the mutation, respectively, which implies that for fibers containing the mutation the length change necessary to drop active force to zero is increased by <10%—i.e., elastic extension of attached cross-bridges (plus myofilaments) is increased by hardly 10%, whereas active force is increased by 59 ± 7% under the same conditions.
Figure 2.
Effect of the Arg719Trp mutation on mechanical parameters of single fibers from soleus muscle. (a) Active isometric force [19 control fibers (open bars) and 18 fibers with mutated myosin (gray bars)] at 10°C. (b) Isometric fiber stiffness [14 control fibers (open bars) and 9 fibers with mutated myosin (gray bars)] at 10°C. (c) Original records of stiffness measurements during releases starting from isometric steady state. Force is plotted vs. change in sarcomere length recorded during the imposed release; speed of releases ≈2.2 × 103 (nm/half-sarcomere) s−1. Upper plot, fiber with Arg719Trp mutation; lower plot, control fiber; solid lines, linear least squares fits to the linear part of the plots (some 3 nm/half-sarcomere for the control fiber and 4 nm/half-sarcomere for the fiber with Arg719Trp mutation). Here, the rather similar intercept of these plots with the x axis (y0 value) shows that approximately the same-length change (release) is necessary to drop active force to zero. Note that, on average, the length change necessary to drop active force to zero was about 10% higher for fibers with mutated myosin than for controls.
Fiber Cross-Sectional Area and Packing of Myofibrils Studied by Confocal Microscopy and X-Ray Diffraction.
Because both active force and fiber stiffness have to be related to fiber cross-section for comparison with normal fibers, the observed differences in force and stiffness critically depend on the precise measurement of the fiber cross-sectional area. For most fibers therefore we recorded optical cross-sections by using a confocal microscope integrated into the mechanical setup to measure the cross-sectional area of the fibers with highest possible precision but without chemical fixation. In addition, these fiber cross-sections together with longitudinal sections allowed evaluation of the packing of the myofibrils in mutant and control fibers. This evaluation was essential because areas without myofibrils in the control fibers could explain the lower force compared with mutant fibers. Fig. 3 a–d shows examples of optical longitudinal and cross-sections through mutant and control fibers labeled with rhodamine-phalloidin. In none of the examined fibers did we find any indication for differences in myofibrillar packing between mutant and control fibers. Finally, to ensure also that the packing of actin and myosin filaments—i.e., the spacing between actin and myosin filaments—and the packing of the myosin filaments themselves are the same in mutant and control fibers, we recorded two-dimensional x-ray diffraction patterns of the fibers. The spacings of the equatorial reflections that correspond to the distance between the thin and thick filaments in the fibers, and of the M3-reflection on the meridian which corresponds to the distance between the crowns of myosin heads along the thick filaments were analyzed. Fig. 3e shows that the spacings of the 1.0 equatorial reflection (d1,0) are essentially the same for mutant and control fibers, both under relaxing and rigor conditions. The spacing of the M3-reflection in rigor was found to be 14.415 ± 0.06 nm (n = 5) for control fibers and 14.421 ± 0.08 nm (n = 8) for mutant fibers, which indicates that the myosin periodicity is the same in both cases. Taken together, these control experiments show that the observed differences in active force and fiber stiffness neither result from uncertainties in the cross-sectional area of the fibers nor from differences in the packing of myofibrils or myofilaments.
Possible Origin of Increased Force and Stiffness.
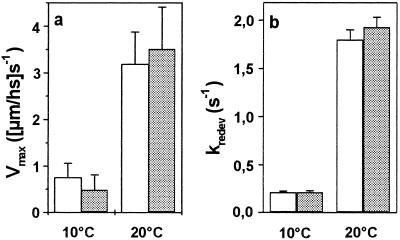
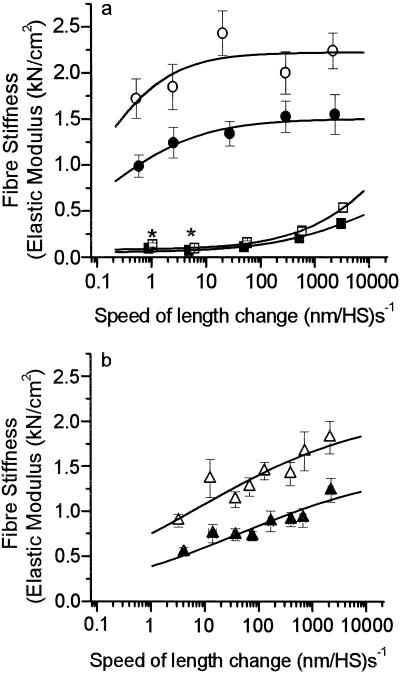
The increase in force and fiber stiffness observed under isometric conditions could result from changes in kinetics of active cross-bridge cycling, such that more cross-bridges occupy force generating states at any one moment (25), or from an increase in force and stiffness of each individual actin-attached cross-bridge (myosin head) (30). We therefore measured the maximum unloaded shortening velocity (vmax) and the rate constant of force redevelopment (kredev) as two parameters sensitive to cross-bridge cycling kinetics. Neither vmax (Fig. 4a) nor kredev (Fig. 4b) were detectably affected by the mutation in the converter domain, neither at 10°C nor at 20°C, indicating that the mutation does not cause a measurable redistribution of the cross-bridges among different intermediate states of the cross-bridge cycle with a higher occupancy of force-generating states under isometric conditions. As for the second possibility, we tested whether the increased force and increased active fiber stiffness result from a higher resistance of the actin-myosin cross-bridges to elastic distortion. Because higher resistance of cross-bridges to elastic distortion might show up in all states of the actomyosin cross-bridge we measured fiber stiffness also in rigor and under relaxing conditions. For a most comprehensive characterization of fiber stiffness, ramp-shaped length changes (stretches) of different speeds were imposed (Fig. 5). In rigor, at the maximum speed of length change, fiber stiffness increased by 45 ± 9% (Fig. 5a). In this condition—i.e., in the absence of nucleotide—essentially all cross-bridges are attached to the actin filament with very high affinity (31, 32), meaning that a stiffness increase in rigor can not be attributed to an increased number of strongly attached myosin heads. Instead, the increase in stiffness of the fibers carrying the mutation in the converter domain must rather be caused by a higher stiffness—i.e., a larger resistance to elastic distortion—of the cross-bridges with the Arg719Trp mutation. Because the mutation changed the elasticity of the myosin heads in rigor (Fig. 5a) and active contraction (Fig. 5b) to a similar extent, it is not surprising that we also found an increased fiber stiffness under relaxing conditions (Fig. 5a)—i.e., in the absence of calcium. Under relaxing conditions myosin heads are weakly attached to actin and can rapidly dissociate during the stiffness measurements (33). Accordingly, only at higher speeds of length change where weakly attached cross-bridges become more and more elastically distorted before they detach, a significant stiffness increase of about 47 ± 8% is detectable. At the lower speeds where passive, non-cross-bridge components dominate, almost no change in fiber stiffness is seen.
Figure 4.
Mechanical parameters sensitive to cross-bridge turnover kinetics. Gray bars, muscle fibers with mutated myosin; open bars, control fibers. (a) Maximum unloaded shortening velocity (vmax) of six fibers each, and (b) rate constant of force redevelopment (kredev) of 21 control fibers and 20 fibers with mutated myosin. Neither vmax nor kredev are affected by the mutation.
Figure 5.
Stiffness of muscle fibers with mutated myosin and of control fibers. (a) Stiffness in rigor (filled circles, 7 control fibers; open circles, 6 fibers with mutation) and under relaxing conditions (filled squares, 14 control fibers; open squares, 10 fibers with mutation), both measured at 5°C. (b) Active isometric stiffness (filled triangles, 14 control fibers; open triangles, 9 fibers with mutation) at 10°C. Lines represent parts of sigmoidal fits to the data points. Stiffness in rigor and relaxation was measured at 5°C to enhance relaxed stiffness. Previous measurements showed an approximately 10% and 15% decrease of relaxed stiffness when raising the temperature from 5°C to 10°C or to 20°C, respectively, whereas rigor stiffness was not affected by these temperature changes. *Difference not statistically significant (P > 0.05).
The observed increase in fiber stiffness and active force, however, cannot be taken as a direct measure of changes in elasticity and force generation of the individual myosin head because the amount of mutated protein in the fibers has to be considered. Because FHC is a dominant autosomal disease, the fibers coexpress wild-type and mutated protein. The ratio of wild-type vs. mutant protein, however, can vary considerably among different mutations (34, 35). Preliminary protein quantification for the Arg719Trp mutation in the fibers of our patient by an approach modified from Nier and coworkers (ref. 35; details will be published elsewhere) revealed that 41 ± 24% (SD, n = 4) of total myosin contains the mutation. This ≈1:1 ratio of mutant to wild-type myosin indicates that individual myosin heads with the mutation are apparently (at least) about twice as stiff and generate about twice as much active force as wild-type myosin heads.
Discussion
Structural Basis of Cross-Bridge Elasticity.
The observed increase in fiber stiffness—i.e., the observed increase in the resistance of fibers to elastic distortion under all conditions studied, including rigor conditions where all cross-bridges are attached to actin—has to be attributed to a stiffer cross-bridge in the presence of the mutation. The location of the mutation in the converter suggests that elastic distortion takes place in the converter domain or in structural elements under strong influence of the converter, and that these structures represent the elastic element of the cross-bridge. A stiffer cross-bridge can only result from increased stiffness of the converter or associated structural elements if other parts of the myosin head/actin complex (cross-bridge) act essentially as rigid bodies—i.e., are much stiffer than the converter—such that most, if not all, of elastic cross-bridge distortion during force generation (or when length changes are imposed on muscle fibers) take place within the converter domain. If, alternatively, essentially all compliance and thus all elastic distortion were located outside the converter (and associated structures), an increased stiffness of the converter (or associated structures) would have essentially no effect on the observed elastic distortion of the cross-bridge. Fiber stiffness would remain unchanged, because observed stiffness is dominated by the element(s) with the smallest resistance to elastic distortion—i.e., the most compliant element(s). Thus, our data suggest that a significant amount of elastic distortion takes place within the converter or associated structures—i.e., structures that are under the influence of the converter such that the Arg719Trp mutation, which is located in the converter, increases their resistance to elastic distortion (stiffness).
This interpretation is consistent with the various crystal structures of the myosin head domain (e.g., refs. 1, 5, 7, 17) in which the converter domain makes rather close contacts with the first three turns of the long α-helix, which forms the core element of the light-chain-binding domain (Fig. 1). As described in detail by Houdusse et al. for scallop striated muscle S1, hydrogen bonds, formed for instance by the conserved Arg-719, but especially clusters of hydrophobic interactions between the first three turns of the long α-helix and elements of the converter domain are most relevant for the structural stability of the converter (17). The stability of the converter, and thus these strong and conserved linkages seem to be essential for transmission of orientational changes in the motor domain by means of the converter to the rest of the light-chain-binding domain helix (17). The point mutation described here directly affects one of the conserved residues of the converter found to interact with the long α-helix (Fig. 1). Although the crystal structure of the head domain of the human β cardiac myosin heavy chain is not known, assuming a structure analogous to those already solved implies that the Arg719Trp mutation increases the hydrophobic interactions and thus could increase the resistance of the converter to elastic distortion when active force is exerted or external forces act on the myosin head. It is possible, however, that the Arg719Trp mutation also affects interactions between residues of the converter domain and nearby residues of the essential light chain (Fig. 1) or, more generally, it might affect the overall structure of the converter domain and its interactions with the light-chain-binding region. By all these routes, the mutation could make the converter itself, the link between converter and light-chain-binding domain, or even the nearby parts of the light-chain-binding domain more resistant to elastic distortion.
Implications of Compliance in the Myofilaments.
About 50% of the total elastic distortion observed in muscle fibers has been reported to take place in the myofilaments (36–38). Such compliance outside the cross-bridges will be distorted more when higher active forces are generated. For instance, with such filament compliance a 50% higher active force in fibers with the Arg719Trp mutation is expected to result in a 25% higher total elastic extension (y0) if the extent of elastic distortion of mutant myosin heads under isometric conditions remains the same as for wild-type myosin heads. Total elastic distortion of mutant fibers during active contraction (y0), however, is only about 10% higher than in control fibers, which can either be accounted for by an elastic distortion of mutant heads (half of all heads) that is only about 40% of the elastic distortion of wild-type heads or if in control fibers elastic extension of myofilaments (myofilament compliance) during isometric force generation is not 50%, but only about 20% of total elastic extension (y0). Our own data suggested that elastic distortion of myofilaments may well be somewhat less than 50% of total elastic distortion during isometric force generation (39). Therefore it seems likely that both factors contribute to the only 10% increase in y0 found here: reduced elastic distortion of mutant myosin heads compared with wild type and somewhat less than 50% myofilament compliance. Thus, elastic distortion of mutant myosin heads is probably not quite as low as only 40% of the wild-type myosin head. If the mutation makes the converter plus associated structures completely rigid, the compliance of the mutant myosin head represents the upper limit of cross-bridge compliance located outside the converter or associated elements.
These estimates illustrate the relevance of filament compliance, which so far we do not know precisely. In addition, the three-dimensional arrangement of cross-bridges and myofilaments in the sarcomere make precise predictions of contributions of cross-bridge and myofilament compliance rather difficult. Thus, for a precise quantitative interpretation other approaches like force and stiffness measurements directly at the single molecule level are needed.
Negative Effects of the Mutation.
Increased active force might lead to the question about why a mutation that is so beneficial for active force generation is causing disease rather than evolving as an advantage. Muscle maximally works with 50% efficiency, and with increased active force of the mutant myosin, one might conclude that the mutation results in an efficiency even higher than 50%. However, to estimate efficiency from work done on elastic elements, we have to distinguish elastic distortion of the attached cross-bridge from elastic extension of myofilaments because the latter does not result from the action of a single head. Thus, only the part of the y0 value that results from elastic distortion of the attached cross-bridge can be considered. This part, however, seems to be significantly reduced for the mutant heads (see above). For example, with 50% of total compliance in the myofilaments of control fibers, 50% higher active force in fibers with the mutation, but only 10% increase in y0, and thus elastic distortion of the individual mutated head reduced to 40% of wild-type myosin, the work done on the elastic elements of the mutant cross-bridge would be 80% the work done on elastic elements of wild-type myosin heads. Thus, the increased active force may actually be associated with reduced efficiency.
Another negative effect that must be considered is the higher load on actin filaments because of the higher force of the mutated cross-bridges. From in vitro force measurements (40) tensile strength of single actin filaments was found to be comparable with the average force exerted on a single thin filament in muscle fibers during isometric contraction. Thus, higher active force might cause rupture of actin filaments resulting in destabilization of the sarcomeres.
Finally, some preliminary results suggest that this mutation also causes a leftward shift of the fore/pCa relation (J.K. and T.K., unpublished results), as seen with other interventions that increase active force (39). This shift is not unexpected if even a small cooperative effect of force-generating cross-bridges on Ca2+ activation exists. However, particularly in cardiac muscle, such a leftward shift of the force-pCa relation would cause impaired relaxation during diastole, which in the Arg719Trp mutation might represent another major disadvantage leading to the typical FHC phenotype and symptoms.
Relation to Other Studies on Mutations in the Converter Domain.
Previous studies with a mutation thought to be equivalent to Arg719Trp introduced either in Dictyostelium myosin II (41) or in smooth muscle myosin (42) were inconsistent in their results and also different from our findings regarding the functional changes caused by the Arg719Trp mutation in cardiac β-MHC. For example, maximum actin-activated ATPase activity in solution, which is thought to correspond to kredev in muscle fibers (25), was found to increase (42) or to decrease (41), whereas in our fiber measurements kredev was not affected by the mutation. Force measured with a laser trap and in vitro sliding velocity were both reduced in the Dictyostelium myosin (41), but were unchanged when the equivalent mutation was introduced in smooth muscle myosin (42). Our measurements of the in vitro sliding velocity (data not shown) and of the maximum shortening velocity in fibers, thought to be related to the in vitro sliding velocity (43, 44), showed no detectable effect of the mutation.
The observed differences between the effects of apparently equivalent mutations in different myosin heavy-chain isoforms indicate that general conclusions about functional consequences of specific FHC mutations derived from experiments on other myosin heavy-chain isoforms must be treated with some caution. Nevertheless, from our work it is expected that an exchange of residues within the converter domain that are in close connection to the first turns of the long α-helix affect elastic properties of the myosin head. Specifically, an increase in the extent of hydrophobic interactions may increase stiffness (i.e., increase the resistance of the myosin head to elastic distortion) as seen here. Identifying the relevant amino acids in myosin heavy-chain isoforms with known crystal structures and mutation to residues with increased/decreased hydrophobicity should enable this hypothesis to be tested.
The significance of the converter domain of the myosin head for the vital functions of the motor molecule is underscored by the fact that patients with this mutation are severely affected by FHC and develop a rather malignant phenotype (16). A whole cluster of mutations is located in the converter domain (14), with the substitutions Arg-723 → Gly and Gly-716 → Arg also resulting in a malignant phenotype (16, 45). These two substitutions are also close to the first turns of the long α-helix. Functional analysis of these and other FHC mutations in the converter domain together with site-directed mutagenesis, expression, and functional analysis of myosin head isoforms with known crystal structure will allow further characterization of the nature of the elastic distortion and testing of our hypothesis that the converter is the main structural element responsible for elastic distortion of the cross-bridge.
Acknowledgments
We are indebted to all individuals who donated their muscle tissue. We thank Dr. A. Berger, Oststadt Krankenhaus, Hannover, and Dr. M. H. J. Becker, Klinikum Wuppertal, Germany, for the muscle biopsies of the control individuals, Birgit Piep, Medical School, Hannover, for excellent technical assistance, and the Forschungswerkstätten of the Medical School for their help. This work was supported by Grant KR 1187/5–1,2 from the Deutsche Forschungsgemeinschaft (to T.K.).
Abbreviations
- β-MHC
β-myosin heavy chain
- FHC
familial hypertrophic cardiomyopathy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rayment I, Holden H M, Whittaker M, Yohn C B, Lorenz M, Holmes K C, Milligan R A. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 2.Lowey S, Waller G S, Trybus K M. Nature (London) 1993;365:454–456. doi: 10.1038/365454a0. [DOI] [PubMed] [Google Scholar]
- 3.Whittaker M, Wilson-Kubalek E M, Smith J E, Faust L, Milligan R A, Sweeney H L. Nature (London) 1995;378:748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- 4.Uyeda T Q, Abramson P D, Spudich J A. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez R, Freyzon Y, Trybus K M, Cohen C. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 6.Corrie J E, Brandmeier B D, Ferguson R E, Trentham D R, Kendrick-Jones J, Hopkins S C, van der Heide U A, Goldman Y E, Sabido-David C, Dale R E, et al. Nature (London) 1999;400:425–430. doi: 10.1038/22704. [DOI] [PubMed] [Google Scholar]
- 7.Geeves M H, Holmes K C. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 8.Huxley A F. Prog. Biophys. Biophys. Chem. 1957. 255–318. [PubMed] [Google Scholar]
- 9.Dobbie I, Linari M, Piazzesi G, Reconditi M, Koubassova N, Ferenczi M A, Lombardi V, Irving M. Nature (London) 1998;396:383–387. doi: 10.1038/24647. [DOI] [PubMed] [Google Scholar]
- 10.Irving M, Piazzesi G, Lucii L, Sun Y B, Harford J J, Dobbie I M, Ferenczi M A, Reconditi M, Lombardi V. Nat Struct Biol. 2000;7:482–485. doi: 10.1038/75890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huxley A F. J Physiol (London) 1974;243:1–43. [PMC free article] [PubMed] [Google Scholar]
- 12.Sugi H, Kobayashi T, Gross T, Noguchi K, Karr T, Harrington W F. Proc Natl Acad Sci USA. 1992;89:6134–6137. doi: 10.1073/pnas.89.13.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisterfer-Lowrance A A, Kass S, Tanigawa G, Vosberg H P, McKenna W, Seidman C E, Seidman J G. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 14.Rayment I, Holden H M, Sellers J R, Fananapazir L, Epstein N D. Proc Natl Acad Sci USA. 1995;92:3864–3868. doi: 10.1073/pnas.92.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonne G, Carrier L, Richard P, Hainque B, Schwartz K. Circ Res. 1998;83:580–593. doi: 10.1161/01.res.83.6.580. [DOI] [PubMed] [Google Scholar]
- 16.Anan R, Greve G, Thierfelder L, Watkins H, McKenna W J, Solomon S, Vecchio C, Shono H, Nakao S, Tanaka H, et al. J Clin Invest. 1994;93:280–285. doi: 10.1172/JCI116957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houdusse A, Kalabokis V N, Himmel D, Szent-Gyorgyi A G, Cohen C. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 18.Houdusse A, Cohen C. Structure (London) 1996;4:21–32. doi: 10.1016/s0969-2126(96)00006-8. [DOI] [PubMed] [Google Scholar]
- 19.Kraft T, Chalovich J M, Yu L C, Brenner B. Biophys J. 1995;68:2404–2418. doi: 10.1016/S0006-3495(95)80423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraft T, Messerli M, Rothen-Rutishauser B, Perriard J C, Wallimann T, Brenner B. Biophys J. 1995;69:1246–1258. doi: 10.1016/S0006-3495(95)80018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruff R L. Muscle Nerve. 1989;12:32–37. doi: 10.1002/mus.880120107. [DOI] [PubMed] [Google Scholar]
- 22.Yu L C, Brenner B. Biophys J. 1989;55:441–453. doi: 10.1016/S0006-3495(89)82838-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner B. Muscle Mechanics II: Skinned Muscle Fibres. London: Oxford Univ. Press; 1998. [Google Scholar]
- 24.Brenner B. Biophys J. 1983;41:99–102. doi: 10.1016/S0006-3495(83)84411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner B, Eisenberg E. Proc Natl Acad Sci USA. 1986;83:3542–6354. doi: 10.1073/pnas.83.10.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill A V. First and Last Experiments in Muscle Mechanics. Cambridge, U.K.: Cambridge Univ. Press; 1970. [Google Scholar]
- 27.Kraft T, Xu S, Brenner B, Yu L C. Biophys J. 1999;76:1494–1513. doi: 10.1016/S0006-3495(99)77309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubis H P, Gros G. Electrophoresis. 1997;18:64–66. doi: 10.1002/elps.1150180113. [DOI] [PubMed] [Google Scholar]
- 29.Ford L E, Huxley A F, Simmons R M. J Physiol (London) 1977;269:441–515. doi: 10.1113/jphysiol.1977.sp011911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner B. Proc Natl Acad Sci USA. 1991;88:10490–10494. doi: 10.1073/pnas.88.23.10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooke R, Franks K. Biochemistry. 1980;19:2265–2269. doi: 10.1021/bi00551a042. [DOI] [PubMed] [Google Scholar]
- 32.Lovell S J, Harrington W F. J Mol Biol. 1981;149:659–674. doi: 10.1016/0022-2836(81)90352-1. [DOI] [PubMed] [Google Scholar]
- 33.Brenner B, Schoenberg M, Chalovich J M, Greene L E, Eisenberg E. Proc Natl Acad Sci USA. 1982;79:7288–7291. doi: 10.1073/pnas.79.23.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malinchik S, Cuda G, Podolsky R J, Horowits R. J Mol Cell Cardiol. 1997;29:667–676. doi: 10.1006/jmcc.1996.0309. [DOI] [PubMed] [Google Scholar]
- 35.Nier V, Schultz I, Brenner B, Forssmann W, Raida M. FEBS Lett. 1999;461:246–252. doi: 10.1016/s0014-5793(99)01433-7. [DOI] [PubMed] [Google Scholar]
- 36.Huxley H E, Stewart A, Sosa H, Irving T. Biophys J. 1994;67:2411–2421. doi: 10.1016/S0006-3495(94)80728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakabayashi K, Sugimoto Y, Tanaka H, Ueno Y, Takezawa Y, Amemiya Y. Biophys J. 1994;67:2422–2435. doi: 10.1016/S0006-3495(94)80729-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higuchi H, Yanagida T, Goldman Y E. Biophys J. 1995;69:1000–1010. doi: 10.1016/S0006-3495(95)79975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraft T, Brenner B. Biophys J. 1997;72:272–281. doi: 10.1016/S0006-3495(97)78666-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kishino A, Yanagida T. Nature (London) 1988;334:74–76. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 41.Fujita H, Sugiura S, Momomura S, Omata M, Sugi H, Sutoh K. J Clin Invest. 1997;99:1010–1015. doi: 10.1172/JCI119228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita H, Tyska M J, Warshaw D M, Lowey S, Trybus K M. J Biol Chem. 2000;275:28045–28052. doi: 10.1074/jbc.M005485200. [DOI] [PubMed] [Google Scholar]
- 43.Homsher E, Wang F, Sellers J. Adv Exp Med Biol. 1993;332:279–289. doi: 10.1007/978-1-4615-2872-2_27. [DOI] [PubMed] [Google Scholar]
- 44.Thedinga E, Karim N, Kraft T, Brenner B. J Muscle Res Cell Motil. 1999;20:785–796. doi: 10.1023/a:1005658825375. [DOI] [PubMed] [Google Scholar]
- 45.Enjuto M, Francino A, Navarro-Lopez F, Viles D, Pare J C, Ballesta A M. J Mol Cell Cardiol. 2000;32:2307–2313. doi: 10.1006/jmcc.2000.1260. [DOI] [PubMed] [Google Scholar]