Abstract
Because the insolubility of the scrapie prion protein (PrPSc) has frustrated structural studies by x-ray crystallography or NMR spectroscopy, we used electron crystallography to characterize the structure of two infectious variants of the prion protein. Isomorphous two-dimensional crystals of the N-terminally truncated PrPSc (PrP 27-30) and a miniprion (PrPSc106) were identified by negative stain electron microscopy. Image processing allowed the extraction of limited structural information to 7 Å resolution. By comparing projection maps of PrP 27-30 and PrPSc106, we visualized the 36-residue internal deletion of the miniprion and localized the N-linked sugars. The dimensions of the monomer and the locations of the deleted segment and sugars were used as constraints in the construction of models for PrPSc. Only models featuring parallel β-helices as the key element could satisfy the constraints. These low-resolution projection maps and models have implications for understanding prion propagation and the pathogenesis of neurodegeneration.
Keywords: electron microscopy‖image processing‖Nanogold labeling‖parallel β-helix‖amyloid structure
Creutzfeldt-Jakob disease (CJD), bovine spongiform encephalopathy (BSE), scrapie, and other spongiform encephalopathies are caused by an aberrantly folded isoform (PrPSc) of the prion protein (PrP) (1). Replication of prions includes a profound change in the conformation of the cellular isoform of PrP (PrPC) to form the highly insoluble PrPSc. The insolubility of PrPSc has thwarted attempts to investigate its structure by either x-ray crystallography or NMR spectroscopy. Our knowledge about the structure of PrPSc is therefore rather limited (2).
After treatment with proteinase K (PK), PrPSc loses the N-terminal residues 23 to ≈89 (forming PrP 27-30), but retains infectivity. During purification, PrP 27-30 polymerizes into rod-shaped filaments with the tinctorial properties of amyloid (3, 4). X-ray fibril diffraction illustrated the amyloid nature of PrP 27-30; characteristic 4.7 Å reflections indicative of cross-β structure were observed (5). Optical spectroscopy revealed that PrPSc and PrP 27-30 are substantially enriched in β-sheet structure (6–9). This finding is in sharp contrast to the predominantly α-helical fold of the three-helix-bundle structure of PrPC as determined by NMR spectroscopy and x-ray crystallography on refolded recombinant PrP (10–18). Owing to the lack of high-resolution structural information for PrPSc, predictive methods have been used to develop molecular models to codify the existing spectroscopic, immunological, and biochemical data (19).
In attempts to simplify the structural analysis of PrPSc, we systematically deleted parts of the prion protein. One of these constructs containing only 106 residues, PrP106 (Δ23–88, Δ141–176), supported the propagation of prions (20, 21). Transgenic mice expressing only PrP106 develop a histologically accurate neurodegenerative prion disease after inoculation with prions, and the resulting prions can be serially passaged (21).
Here we report the discovery of two-dimensional (2D) crystals of PrP 27-30 and PrPSc106. Electron micrographs of the crystals were analyzed by digital image processing and found to have three-fold symmetry. The complexation of heavy metal cations onto the crystal lattice indicated the presence of a strong negative electrostatic potential in the center of individual oligomers. Specific labeling of the N-linked sugars with Monoamino Nanogold (Nanoprobes, Yaphank, NY) enabled us to localize them toward the outside of the oligomer. Crystals of PrPSc106, isomorphous to the crystals of PrP 27-30, permitted the determination of significant differences between the two structures. Difference mapping revealed the location of the 36-residue internal deletion of the miniprion in projection. These data were coupled with a variety of other experimental results to create constraints on plausible models for the structure of PrPSc. Only models featuring a parallel β-helix as the key element could satisfy all of these constraints.
Materials and Methods
Negative Stain Electron Microscopy.
The negative staining and electron microscopy (EM) were performed as described (22). Electron micrographs used for image processing were recorded at a magnification of 80,000 and an electron dose of ≈25 pA/cm2 (equivalent to ≈100 electrons/Å2).
Image Processing.
Negatives of suitable 2D crystals were scanned on a Perkin–Elmer PDS microdensitometer with a pixel size of 20 μm (equivalent to 2.5 Å per pixel). A contrast transfer function (CTF) correction was applied by using the crisp software package (23). In some cases, crisp was also used to partially compensate for the dampening of the CTF function toward higher resolution. The CTF-corrected images were processed by using correlation-mapping and averaging routines written for the spider and web software package (24). A manually chosen 128 × 128 pixel reference was cross-correlated to same-size subsets of the image in a rotational and translational search. After multiple iterations, the algorithm converged and no further improvements could be achieved. The aligned subsets of the CTF-corrected micrograph were examined by correspondence analysis (24) to ensure that only a homogeneous population of images was used. Finally, the data of the original electron micrograph were divided into 10 sectors and averaged separately. These 10 averages were then used for plane group analysis and crystallographic averaging. The 10 crystallographic subaverages were combined into one final average.
Labeling of the N-Linked Sugars.
The labeling procedure was modified after Lipka et al. (25). Suspensions of PrP 27-30 2D crystals were oxidized (10 mM NaIO4/100 mM NaHepes, pH 7.0) for 2 h at room temperature (RT). The protein was kept in suspension by end-over-end rotating. After the oxidation, the protein was pelleted and resuspended in carbonate buffer (100 mM sodium carbonate, pH 9.0). An excess of 1.4 nm Monoamino Nanogold (Nanoprobes) was suspended in carbonate buffer and added to the protein. The oxidized 2D crystals and the Monoamino Nanogold were reacted for 2 h at RT under constant rotation. The reaction was brought to completion by adding a small aliquot of 5 M NaBH4 in 0.1 M NaOH. This reaction was allowed to sit undisturbed for 30 min at RT. Finally, the protein was pelleted, the unbound gold label was discarded with the supernatant, and the pellet was resuspended in buffer. Controls were treated identically with the exception that no Monoamino Nanogold was added.
The sugar residues within the glycosylphosphatidylinositol (GPI) anchor made it necessary to establish that the label was indeed bound to the N-linked sugars. We denatured gold-labeled PrP 27-30 and treated it with PNGase F. Western blots of unlabeled, labeled, and enzyme-treated samples were developed with the 3F4 Ab or a silver enhancement kit that directly visualized the gold label on the blotting membrane. The labeled samples showed an additional band at ≈45 kDa that was detected by both 3F4 and the silver enhancement, indicating that this band corresponded to the gold-labeled PrP 27-30 (data not shown). Treatment with PNGase F removed the gold label, thus showing that the Nanogold label indeed specifically bound to the N-linked sugars.
Image Processing of Labeled Crystals.
The image processing routine, as developed for unlabeled 2D crystals, tended to cancel out the signal of the Nanogold particles because the alignment procedure favored lattice-derived signals over the somewhat irregular gold labels. After the iterative alignment procedure, we used correspondence analysis (24) to eliminate the contributions of unlabeled subunits. Furthermore, we separated four different classes of partially labeled subunits, calculated the average for each class, and then used those averages for subsequent crystallographic averaging.
Modeling.
Models were built by using swiss-pdb-viewer software (26). Briefly, the PrP structure was threaded onto known β-helical structures in all possible registers, and models were built with the helices packed against all possible sides. The models that provided the most stericly reasonable faces for assembly and electrostatic surfaces that most closely resembled the observed negative staining were selected. In addition, an effort was made to preserve the location of secondary structural elements predicted by a variety of methods (27–29) or indicated by experimental observation. Figures were created with swiss-pdb-viewer and rasmol.
Results
2D Crystals of PrP 27-30.
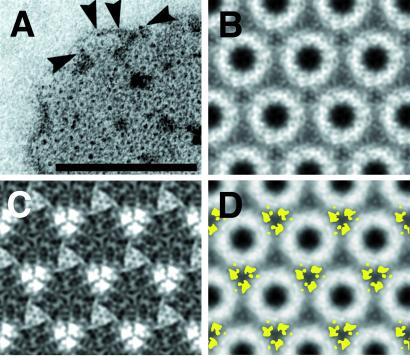
Negative stain EM revealed the presence of 2D protein crystals in some preparations of PrP 27-30 (Fig. 1A). During the purification of PrP 27-30, most of the protein polymerized into “prion rods,” typical amyloid polymers that revealed little structural detail (3, 4). The final step of this purification procedure used a sucrose gradient centrifugation (3). We discovered that some fractions contain prion rods and 2D crystals (Fig. 1A) with an apparent hexagonal lattice (a and b = 69 Å; γ = 120° as determined by electron diffraction).
Figure 1.
2D crystals of PrP 27-30. (A) A 2D crystal of PrP 27-30 stained with 2% uranyl acetate showing an apparent hexagonal lattice. (B) High power view of a crystal after CTF correction and several rounds of correlation-mapping and averaging. (C) Section of a power spectrum after averaging showing spots out to the 11th order, corresponding to ≈7 Å (arrow). (D) Crystallographic averaging further improved the amount of detail visible. A p3 plane group was used. (E) Typical prion rod with an aggregate of “crystal” subunits at each end. Some protofilaments reveal rows of dense stain accumulations, suggesting stacked subunits (arrowheads). [Bars = 100 nm.]
Immunogold labeling with anti-PrP mAbs R1, R2, 3F4, and 28D established that PrP is an integral part of these crystals (data not shown). Decoration with 3F4 could be achieved only after urea denaturation, arguing that the scrapie isoform is present in these crystals, because 3F4 recognizes an epitope that is inaccessible in native PrPSc (30, 31). Furthermore, the 2D crystals originated in preparations that had high titers of infectivity and had not been treated by denaturing agents. Therefore, we consider the crystals to be fully infectious. Moreover, we frequently observed 2D crystals in the direct vicinity of the prion rods and on occasion what appeared to be transitions between them (Fig. 1E). These transitional aggregates suggest a stacking of the disk-like oligomers into “protofilaments.”
The 2D crystals can be visualized with negative stains such as uranyl acetate (Fig. 1A). Various heavy metal stains, including other uranyl salts, cerium chloride, and ammonium molybdate stained the crystals equally well (data not shown). We reported on 2D crystals of PrP 27-30 that were obtained from reverse micelles but that lacked infectivity (32). Unlike the crystals described here, those noninfectious crystals strictly depended on divalent uranyl cations for crystallization.
The dark areas in the center of the current 2D crystal subunits (Fig. 1 A and E) are caused by a complexation of uranyl ions (or other heavy metal cations) to negative charges within the crystal lattice, effecting a positive stain. The nature of the complex formation became apparent when uranyl salts that differ greatly in their stability constants (pK) were used. For example, uranyl acetate and oxalate produced the typical staining pattern seen in Fig. 1A. In contrast, uranyl citrate generated a very different result with no apparent staining of the subunit centers (data not shown). The pK values for uranyl acetate, oxalate, and citrate are 3.2, 6.5, and 18.9, respectively (33). Thus, we know that the uranyl⋅PrP 27-30 complex has an effective pK greater than 6.5 but less than 18.9. The observation that anionic stains, like molybdate, were repelled from the center of the subunits (data not shown) allowed us to conclude that a cluster of partial or full negative charges occupies the center of each oligomer. The sequence of PrP 27-30 contains several negatively charged residues that may contribute to the observed complexation of heavy metal cations; the highest concentration of these residues lies between amino acids 143 and 177.
Image Processing.
To obtain images suitable for digital processing, we took electron micrographs of the 2D crystals at underfocus values lower than those of conventional negative stain images. The high contrast of the negatively stained crystals enabled us to process images with 200–300 nm of underfocus, as determined from the Thon rings of Fourier transforms. Because of the limited quality and small size of the crystals, the images were processed with correlation methods that are used routinely for single-particle reconstructions. A region containing several unit cells was correlated throughout the crystal, and appropriate rotations and translations were applied to correct for local lattice imperfections. Fig. 1B shows the results after several rounds of iterative correlation-mapping and averaging. The power spectrum showed spots out to the 11th order, which corresponds to ≈7 Å spacing (Fig. 1C).
Crystallographic averaging exploits the symmetry relationships within the Fourier transforms of individual images. The amplitude R factors and phase residuals for a 2D crystal processed for the relevant plane groups indicate a p3 plane group as the most likely symmetry (Table 1). Thus, we routinely applied p3 symmetry during crystallographic averaging (Fig. 1D). Because rotational cross-correlation demonstrated six distinct densities within each subunit (data not shown), we presuppose either a trimer of dimers arrangement for the unit cell with a noncrystallographic dimer axis or a trimeric arrangement, in which each subunit has two centers of density. The trimeric arrangement would give the unit cell a polarity that should be noticeable within the crystals and could result in a polar filament assembly. As neither of these features was observed at the current resolution, we favor the trimer of dimers interpretation.
Table 1.
Plane group statistics for PrP 27-30 electron micrograph no. 12822
| Plane group | Amplitude Rsym factor, %
|
Amplitude-weighted phase residual, °
|
||
|---|---|---|---|---|
| Individual* | (Average)† | Individual* | (Average)† | |
| p1 | — | — | — | — |
| p21 | — | — | 22.6 | (24.7) |
| p3 | 9.1 | (10.9) | 10.9 | (15.8) |
| p312 | 11.1 | (15.0) | 19.1 | (26.2) |
| p321 | 11.1 | (15.0) | 18.5 | (25.7) |
| p6 | 9.1 | (10.9) | 27.6 | (29.6) |
| p622 | 11.1 | (15.0) | 26.9 | (33.4) |
Image divided into 10 sectors that were averaged and analyzed independently.
Data for the image sector that allowed the clearest distinction between different plane groups.
Average of the R factors and phase residuals for all 10 sectors.
Localization of the Sugar Side Chains.
To localize the N-linked sugars of PrP 27-30, we specifically labeled the oligosaccharides by 1.4-nm gold particles (Nanogold). The procedure succeeded in labeling the preformed crystals without affecting prion infectivity (data not shown).
The high contrast of uranyl acetate staining made it difficult to visualize the small label (Fig. 2A). The image processing on labeled crystals (Fig. 2 A and B) revealed some differences, but the procedure required additional refinements to detect the somewhat nonperiodic labels. Because the crystallographic averaging used p3 symmetry, the procedure may have systematically underestimated the number of locations for the gold label by a factor of 2. The subtraction map did indeed show some weak differences at the second three-fold axis (Fig. 2C). By subtracting 3 times the value of the standard error map, we obtained statistically significant differences that when overlaid onto the average of an unlabeled crystal, showed the sugar side chains located toward the outside of the oligomers (Fig. 2D). This peripheral localization provides a constraint on the orientation of the protein molecules within the crystal lattice.
Figure 2.
Nanogold labeling of the N-linked sugars. (A) Uranyl acetate-stained 2D crystal of Nanogold-labeled PrP 27-30. The high contrast of the uranyl stain obscures some of the labels, but others are clearly visible (arrowheads). [Bar = 100 nm.] (B) Image processing result of a labeled crystal after correlation-mapping and averaging followed by crystallographic averaging. (C) Subtraction map between labeled and unlabeled crystals showing major differences in lighter shades. (D) Overlay of the statistically significant differences calculated from C in yellow onto a projection map of PrP 27-30.
The N-linked sugars of hamster PrP 27-30 are bound to residues N181 and N197. In the solution structure of recombinant PrPC, these residues lie in helix B and near the N terminus of helix C, respectively (10–17). Theoretical considerations and experimental evidence indicate that helices B (residues 179–193) and C (residues 200–217) in PrPC are preserved in PrPSc (19). Fourier transform infrared spectroscopy of PrP 27-30 and PrPSc106 demonstrated substantial residual α-helical structure (23% and 27–31%, respectively) (21, 22), whereas epitope mapping localized the conformational change that characterizes PrPSc to the region between residues 90 and 170 (34). Synthetic peptides corresponding to PrP(90–145,P101L) have been shown to refold into a β-sheet-rich state that can initiate or accelerate prion disease in specific transgenic mice (35), reinforcing the notion that the formation of β-structure in PrPSc is unlikely to involve the C-terminal helices. Thus, residues N181 and N197 are expected to be closely linked to the remaining helical regions of PrPSc. As these segments are joined by a disulfide bridge (C179-C214) that is retained in PrPSc and required for infectivity (20), it is likely that the α-helices of PrPSc will be located toward the outside of the oligomers as well.
The relatively small lattice parameters and the accordingly dense packing of the protein molecules in the trimer of dimers arrangement would require the sugars to protrude up and down from the lattice. The projection map alone does not allow us to confirm whether the sugars are in or out of the plane of the protein moiety. The less dense packing of a trimeric assembly would give the sugars additional room in the lattice. The intrinsic flexibility of the sugar side chains of PrP may allow them to rotate in and out of the lattice plane, depending on steric constraints (36). This notion is supported by the observation that Nanogold-labeled prion rods show a relatively dense covering of gold labels on the surface of the rods (data not shown). This result in turn suggests that the lateral association of “protofilaments” into prion rods is influenced—if not dominated—by the N-linked sugars. Furthermore, steric hindrance by the oligosaccharides could account for the inaccessibility of prion rods to various enzymatic digestions.
Differences Between PrP 27-30 and PrPSc106.
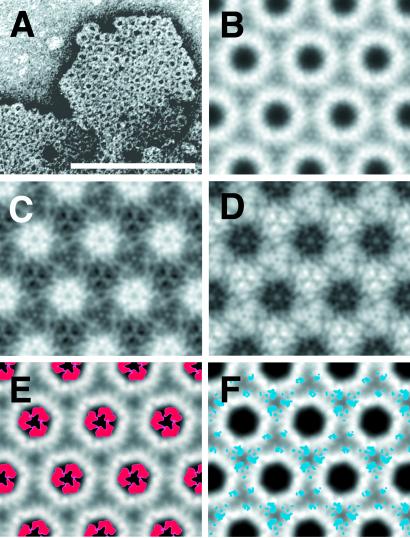
While analyzing preparations of PrPSc106 miniprions by negative stain EM, we observed both rod-like polymers (21) and 2D crystals isomorphous to those seen with PrP 27-30 (Fig. 3A). This finding enabled us to use the same image processing strategies used for the PrP 27-30 crystals (Figs. 1D and 3B), and map the differences between PrP 27-30 and PrPSc106.
Figure 3.
2D crystals of PrPSc106. (A) A 2D crystal of PrPSc106 stained with uranyl acetate. [Bar = 100 nm.] (B) Image processing result after correlation-mapping and averaging followed by crystallographic averaging. (C and D) Subtraction maps between the averages of PrP 27-30 (Fig. 1D) and PrPSc106 (B). (C) PrPSc106 minus PrP 27-30 and (D) PrP 27-30 minus PrPSc106, showing major differences in lighter shades. (E and F) The statistically significant differences between PrP 27-30 and PrPSc106 calculated from C and D in red and blue, respectively, overlaid onto the crystallographic average of PrP 27-30 (Fig. 1D).
PrPSc106 lacks residues 141–176 but is consistently diglycosylated (20, 21). By comparison, PrP 27-30 has a longer peptide chain and is a mix of un-, mono-, and diglycosylated forms (36). As these crystals display a complex mixture of negative and positive staining, it is important to consider the staining behavior of the different constructs. PrPSc106 lacks many of the negatively charged residues that are present in PrP 27-30 and hence binds fewer uranyl cations at the center of the subunits (data not shown). Therefore, subtracting the PrP 27-30 density from the PrPSc106 density should highlight residues 141–176 in positive density, whereas subtracting PrPSc106 from PrP 27-30 should place the extra glycosylation sites in positive density.
As expected, PrP 27-30 minus PrPSc106 showed differences that were nearly identical to the location of the sugars determined by Nanogold labeling (Figs. 2D and 3F). Here the difference signal for the N-linked sugars was found on all three-fold symmetry axes around the oligomer (Fig. 3F). PrPSc106 minus PrP 27-30 revealed differences around the center of each oligomer (Fig. 3E), which we interpret as being representative of the amino acids in the internal deletion of PrP106 (Δ141–176).
Fourier transform infrared spectroscopy indicates a β-sheet content of ≈48% for PrP 27-30 (≈68 of 142 residues) and ≈37% for PrPSc106 (≈39 of 106 residues) when measured under identical conditions (21, 22). By comparing these percentages, we conclude that most of the 36 residues of the internal deletion adopt a β-sheet conformation in the scrapie isoform. We therefore infer that the difference between PrP 27-30 and PrPSc106 visualized in our 2D crystals (Fig. 3E) actually represents predominantly β-sheet structure.
In the solution structure of recombinant PrPC, residues 141 and 176 are separated by ≈18 Å with helix C physically intervening between the two residues (10–17). That PrP 27-30 and PrPSc106 form isomorphous 2D crystals requires residues 140 and 177 to be in close proximity to each other in the scrapie conformation. Thus, a two- or four-stranded β-sheet motif with spatially proximal termini like a β-hairpin, a β-meander, a Greek key motif, or two turns of a parallel β-helix are plausible structural arrangements.
Models of PrPSc.
Previous endeavors to model the structure of PrPSc attempted to fit sequence-specific conformational preferences with spectroscopic, antibody-binding, and other biological data. Originally, we postulated an anti-parallel β-sheet formed from residues 90 to 170 packed against the two C-terminal α-helices (19). The results obtained from the 2D crystals described here, the existence of PrPSc106 (implying the spatial colocalization of residues 140 and 177), and increasing data pointing to parallel β-sheet structure in amyloid-forming proteins (37, 38) caused us to revisit this model (19).
A single anti-parallel β-sheet was not consistent with the observed densities in the projection maps obtained from the 2D crystals. Specifically, the sheets were far too wide to fit into the observed hexameric arrangement. Efforts to adjust the sheet morphology to fit the density required the use of shorter strands. The amount of β-structure in these altered β-sheets was no longer compatible with the amounts of β-sheet observed by Fourier transform infrared spectroscopy (21, 22). Furthermore, anti-parallel β-sheets typically have a twist of ≈20° per strand. A six- to eight-stranded β-sheet would be difficult to accommodate in the electron density. Parallel β-sheets are commonly observed in protein structures as part of planar α/β folds, α/β barrels, and parallel β-helices. Planar α/β folds encounter the same problems as twisted anti-parallel β-sheets. α/β barrels have alternating α-helices and β-strands with the fraction of α-helical residues exceeding that of the β-stranded residues. The secondary structure content of these folds would be in conflict with the FTIR results for both PrP 27-30 and PrPSc106. Although we cannot exclude a novel protein fold for the structure of PrPSc, the parallel β-helix is the only known fold that provides the necessary β-sheet content, parallel β-architecture, and room to accommodate the α-helices that are expected at the C-terminus of the molecule.
A number of proteins natively form parallel β-helices and exist as soluble monomers or low-order oligomers (39–42). This fold is considered unusually stable, and mutants of these proteins have been observed to form amyloid (43). There are essentially two types of parallel β-helical folds: the left- and right-handed β-helices. Proteins of either type are known to contain highly ordered, stacked side chains on both the inside and outside of the β-helices (41). This side-chain stacking adds to the rigid geometry of the β-helices, which tend to have planar sides and essentially no interstrand twist (41). The number of residues per β-helical turn can vary substantially for right-handed β-helices (39, 41, 42), whereas an average of 18 residues per helical turn is found in the known left-handed β-helical structures (40).
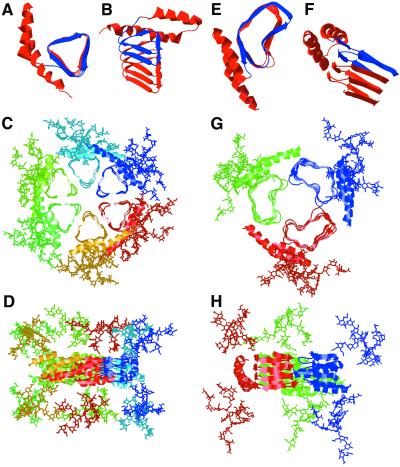
We modeled PrPSc as a parallel β-helical fold (Fig. 4 A and E), placing the structurally conserved C-terminal α-helices and the glycosylation sites (N181 and N197) on the periphery of the oligomer and with the highly flexible N-linked sugars pointing above and below the plane of the oligomer (Fig. 4 D and H). In this conformation, PrPSc is compact (Fig. 4 C and G) and fits readily into the density observed by EM. Because there is very little twist or bend to parallel β-helices, the modeled oligomers have relatively planar faces that permit stacking along the fibril axis. The β-helices also provide flat sheets for lateral assembly into disk-like oligomers and filamentous assemblies (42, 43). The deletion of 36 residues in PrP106 correlates favorably with exactly two turns of an average left-handed β-helix. Therefore, the orientation of the α- and β-helices, the sugars, as well as the fold of the oligomeric face, could be retained in this deletion mutant. This would allow full-length PrPSc as well as PrPSc106 to template the replication of PrPSc106. Furthermore, the shape and location of the β-helical deletions in our models are consistent with the difference densities observed between the 2D crystals of PrP 27-30 and PrPSc106. Finally, the PrP sequence can be threaded onto the β-helical folds in a register that is consistent with secondary structure predictions, mutational information, and the negative electrostatic potential at the center of the oligomers.
Figure 4.
β-Helical models of PrP 27-30. (A and B) Top and side views, respectively, of PrP 27-30 modeled with a left-handed β-helix. The β-helical portion of the model is based on the Methanosarcina thermophila γ-carbonic anhydrase structure. (C and D) Top and side views, respectively, of the trimer of dimer model of PrP 27-30 with left-handed β-helices. (E and F) Top and side views, respectively, of PrP 27-30 modeled with a right-handed β-helix. The β-helical portion of the model is based on the most regular helical turns of Bordetella pertussis P.69 pertactin. (G and H) Top and side views, respectively, of the trimer model of PrP 27-30 with right-handed β-helices. The structure of the α-helices was derived from the solution structure of recombinant hamster PrP (11–13). In the single-molecule images (A, B, E, and F), residues 141–176 that are deleted in PrP106 are colored blue.
Discussion
We report the discovery of 2D crystals in purified infectious fractions of PrP 27-30 and PrPSc106 by negative stain EM. The 2D crystals were found exclusively in preparations that contain high titers of prion infectivity. Because crystals were found in close proximity to prion rods (Fig. 1E) and the Ab 3F4 recognized them only after urea treatment (data not shown), we conclude that the crystals contain the infectious isoform of the prion protein.
Power spectra of digitally processed images showed spots out to the 11th order at ≈7 Å (Fig. 1C). Because the micrographs were taken with high doses of electrons, we were surprised to obtain such high-resolution data. Given that the stained crystals bound heavy metal cations in the center of their subunits, we suspect that the high-resolution information is limited to those areas. In addition, the presence of heavy metal cations may have provided some partial protection against irradiation damage. The current data are limited to projection maps only; a three-dimensional reconstruction will give more detailed insight into the structure of PrPSc. The future use of low-dose cryoelectron crystallography may allow us to solve the structure of this heretofore intractable isoform.
The observed crystals were obtained from preparations of different prion strains and PrP constructs: Syrian hamster (SHa) Sc237 prions (Fig. 2; Table 1), mouse (Mo) RML prions (Fig. 1), and chimeric mouse–hamster (MHM2) RML-PrP106 prions (Fig. 3 A and B). Prion strains seem to be enciphered in the conformation of PrPSc and domain swapping has been offered as a structural explanation for the existence of several isoenergetic PrPSc structures (31, 44–49). A direct comparison of the Sc237 and RML prions used in this study is difficult owing to the eight-residue differences between the hamster and mouse sequences.
In summary, we report that PrP 27-30 has the ability to form 2D crystals, the subunits of which expose a cluster of negative charges at their center that are involved in the complexation of heavy metal cations. By image subtraction, we were able to localize the N-linked sugars of PrP 27-30 toward the periphery of the crystal subunits. Additionally, internally deleted PrPSc106 forms 2D crystals that are isomorphous to those of PrP 27-30; difference mapping between PrP 27-30 and PrPSc106 revealed the location of the 36-residue internal deletion at the inside of the oligomer. Optical spectroscopy suggested that these 36 residues are predominantly in a β-sheet conformation (21, 22). The isomorphism of the PrP 27-30 and PrPSc106 crystals implies a close proximity between residues 140 and 177 in both proteins, consistent with a β-sheet-rich fold such as a parallel β-helix. By combining these new constraints with other experimental data, we can exclude many structural motifs and argue that PrPSc is likely to contain a parallel β-helix.
The parallel β-helix that we propose for PrPSc (Fig. 4) is considered an unusually stable fold, and is generally found in proteins subjected to harsh, denaturing environments such as bacterial or viral virulence factors or plant pollens (41). Furthermore, the fold is very simple and may form in a two-state manner analogous to α-helix formation (42). Thus, the conversion from PrPC to PrPSc can be understood as a stabilization of a proto-β-helical motif by a neighboring PrPSc molecule and subsequent extension to form the complete β-helix.
Acknowledgments
We thank Robert M. Stroud, Zoltan F. Kanyo, Vinzenz Unger, and Sven Hovmöller for their advice on crystallography and computational issues; Sebastian Doniach for suggesting the trimer of dimers arrangement; John M. Chandonia for assistance with structural threading; Pauline M. Rudd for discussions on the glycosylphosphatidylinositol-anchor and N-linked sugars; Susanne D. Erpel and Diane Latawiec for expert technical assistance; Kenneth H. Downing for access to the microdensitometer; and Mei-Lie Wong for making darkroom facilities available. M.D.M. acknowledges a National Science Foundation predoctoral fellowship. V.G. was supported by a Howard Hughes postdoctoral fellowship. S.S. was supported by a Burroughs Wellcome Fund Career Development Award and a National Institutes of Health Clinical Investigator Development Award. This work was supported by grants from the National Institutes of Health and a gift from the G. Harold and Leila Y. Mathers Charitable Foundation.
Abbreviations
- PrP
prion protein
- PrPC
normal cellular isoform
- PrPSc
disease-associated isoform
- PrP 27-30
N-terminally truncated PrPSc
- 2D
two-dimensional
- EM
electron microscopy
- CTF
contrast transfer function
References
- 1.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen F E, Prusiner S B. In: Prion Biology and Diseases. Prusiner S B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 191–228. [Google Scholar]
- 3.Prusiner S B, McKinley M P, Bowman K A, Bolton D C, Bendheim P E, Groth D F, Glenner G G. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 4.McKinley M P, Meyer R K, Kenaga L, Rahbar F, Cotter R, Serban A, Prusiner S B. J Virol. 1991;65:1340–1351. doi: 10.1128/jvi.65.3.1340-1351.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen J T, Inouye H, Baldwin M A, Fletterick R J, Cohen F E, Prusiner S B, Kirschner D A. J Mol Biol. 1995;252:412–422. doi: 10.1006/jmbi.1995.0507. [DOI] [PubMed] [Google Scholar]
- 6.Caughey B W, Dong A, Bhat K S, Ernst D, Hayes S F, Caughey W S. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 7.Gasset M, Baldwin M A, Fletterick R J, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:1–5. doi: 10.1073/pnas.90.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safar J, Roller P P, Gajdusek D C, Gibbs C J., Jr J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 10.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 11.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E, Dyson H J. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Farr-Jones S, Ulyanov N B, Llinas M, Marqusee S, Groth D, Cohen F E, Prusiner S B, James T L. Biochemistry. 1999;38:5362–5377. doi: 10.1021/bi982878x. [DOI] [PubMed] [Google Scholar]
- 14.Calzolai L, Lysek D A, Güntert P, von Schroetter C, Riek R, Zahn R, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:8340–8345. doi: 10.1073/pnas.97.15.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García F L, Zahn R, Riek R, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:8334–8339. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, López García F, Billeter M, Calzolai L, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Swietnicki W, Zagorski M G, Surewicz W K, Sönnichsen F D. J Biol Chem. 2000;275:33650–33654. doi: 10.1074/jbc.C000483200. [DOI] [PubMed] [Google Scholar]
- 18.Knaus K J, Morillas M, Swietnicki W, Malone M, Surewicz W K, Yee V C. Nat Struct Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 19.Huang Z, Prusiner S B, Cohen F E. Folding Des. 1995;1:13–19. [PubMed] [Google Scholar]
- 20.Muramoto T, Scott M, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1996;93:15457–15462. doi: 10.1073/pnas.93.26.15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Supattapone S, Bosque P, Muramoto T, Wille H, Aagaard C, Peretz D, Nguyen H-O B, Heinrich C, Torchia M, Safar J, et al. Cell. 1999;96:869–878. doi: 10.1016/s0092-8674(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 22.Wille H, Zhang G-F, Baldwin M A, Cohen F E, Prusiner S B. J Mol Biol. 1996;259:608–621. doi: 10.1006/jmbi.1996.0343. [DOI] [PubMed] [Google Scholar]
- 23.Hovmöller S. Ultramicroscopy. 1992;41:121–135. [Google Scholar]
- 24.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 25.Lipka J J, Hainfeld J F, Wall J S. J Ultrastruct Res. 1983;84:120–129. doi: 10.1016/s0022-5320(83)90123-5. [DOI] [PubMed] [Google Scholar]
- 26.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 27.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 28.Garnier J, Gibrat J F, Robson B. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 29.Chandonia J M, Karplus M. Proteins. 1999;35:293–306. [PubMed] [Google Scholar]
- 30.Peretz D, Williamson R A, Matsunaga Y, Serban H, Pinilla C, Bastidas R B, Rozenshteyn R, James T L, Houghten R A, Cohen F E, et al. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 31.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 32.Wille H, Prusiner S B. Biophys J. 1999;76:1048–1062. doi: 10.1016/S0006-3495(99)77270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martell A E, Smith R M. Critical Stability Constants. New York: Plenum; 1974. [Google Scholar]
- 34.Williamson R A, Peretz D, Pinilla C, Ball H, Bastidas R B, Rozenshteyn R, Houghten R A, Prusiner S B, Burton D R. J Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko K, Ball H L, Wille H, Zhang H, Groth D, Torchia M, Tremblay P, Safar J, Prusiner S B, DeArmond S J, Baldwin M A, Cohen F E. J Mol Biol. 2000;295:997–1007. doi: 10.1006/jmbi.1999.3386. [DOI] [PubMed] [Google Scholar]
- 36.Rudd P M, Wormald M R, Wing D R, Prusiner S B, Dwek R A. Biochemistry. 2001;40:3759–3766. doi: 10.1021/bi002625f. [DOI] [PubMed] [Google Scholar]
- 37.Benzinger T L S, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Proc Natl Acad Sci. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antzutkin O N, Balbach J J, Leapman R D, Rizzo N W, Reed J, Tycko R. Proc Natl Acad Sci. 2000;97:13045–13050. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoder M D, Keen N T, Jurnak F. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 40.Raetz C R, Roderick S L. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 41.Jenkins J, Mayans O, Pickersgill R. J Struct Biol. 1998;122:236–246. doi: 10.1006/jsbi.1998.3985. [DOI] [PubMed] [Google Scholar]
- 42.Seckler R. J Struct Biol. 1998;122:216–222. doi: 10.1006/jsbi.1998.3974. [DOI] [PubMed] [Google Scholar]
- 43.Schuler B, Rachel R, Seckler R. J Biol Chem. 1999;274:18589–18596. doi: 10.1074/jbc.274.26.18589. [DOI] [PubMed] [Google Scholar]
- 44.Bessen R A, Marsh R F. J Virol. 1994;68:7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telling G C, Parchi P, DeArmond S J, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner S B. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 46.Scott M R, Groth D, Tatzelt J, Torchia M, Tremblay P, DeArmond S J, Prusiner S B. J Virol. 1997;71:9032–9044. doi: 10.1128/jvi.71.12.9032-9044.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen F E, Prusiner S B. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 48.Wadsworth J D F, Hill A F, Joiner S, Jackson G S, Clarke A R, Collinge J. Nat Cell Biol. 1999;1:55–59. doi: 10.1038/9030. [DOI] [PubMed] [Google Scholar]
- 49.Peretz D, Scott M, Groth D, Williamson A, Burton D, Cohen F E, Prusiner S B. Protein Sci. 2001;10:854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]