Abstract
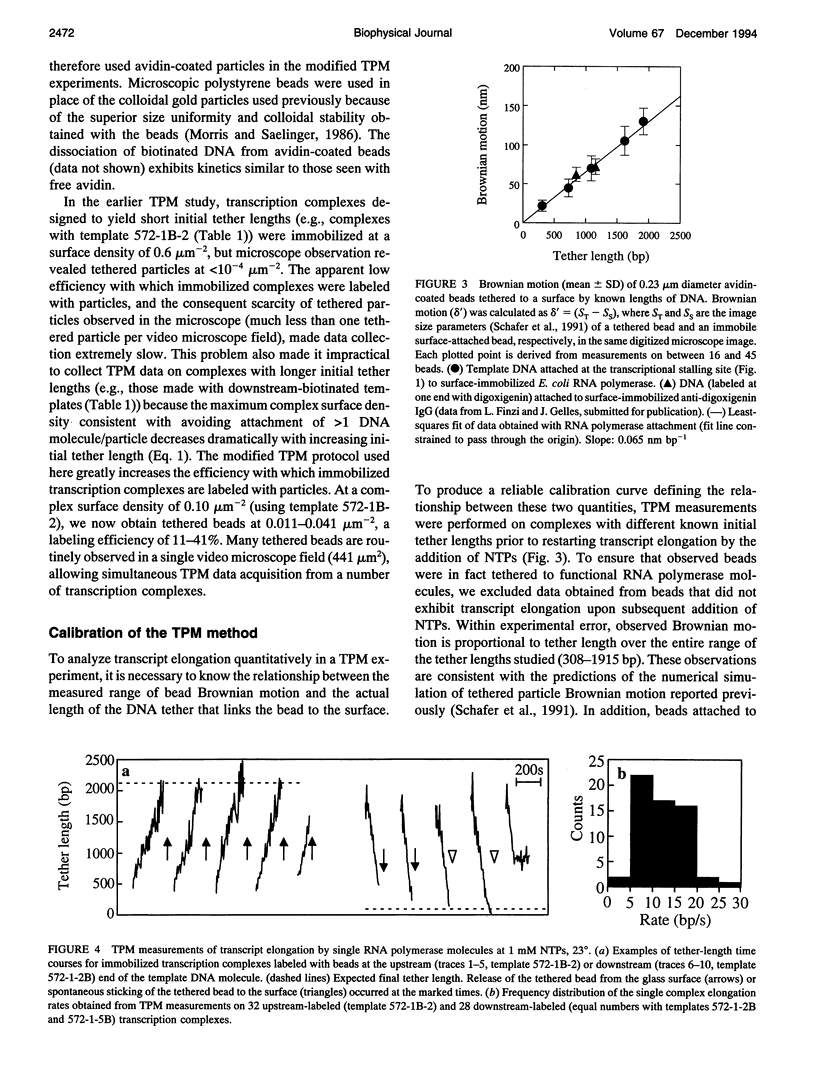
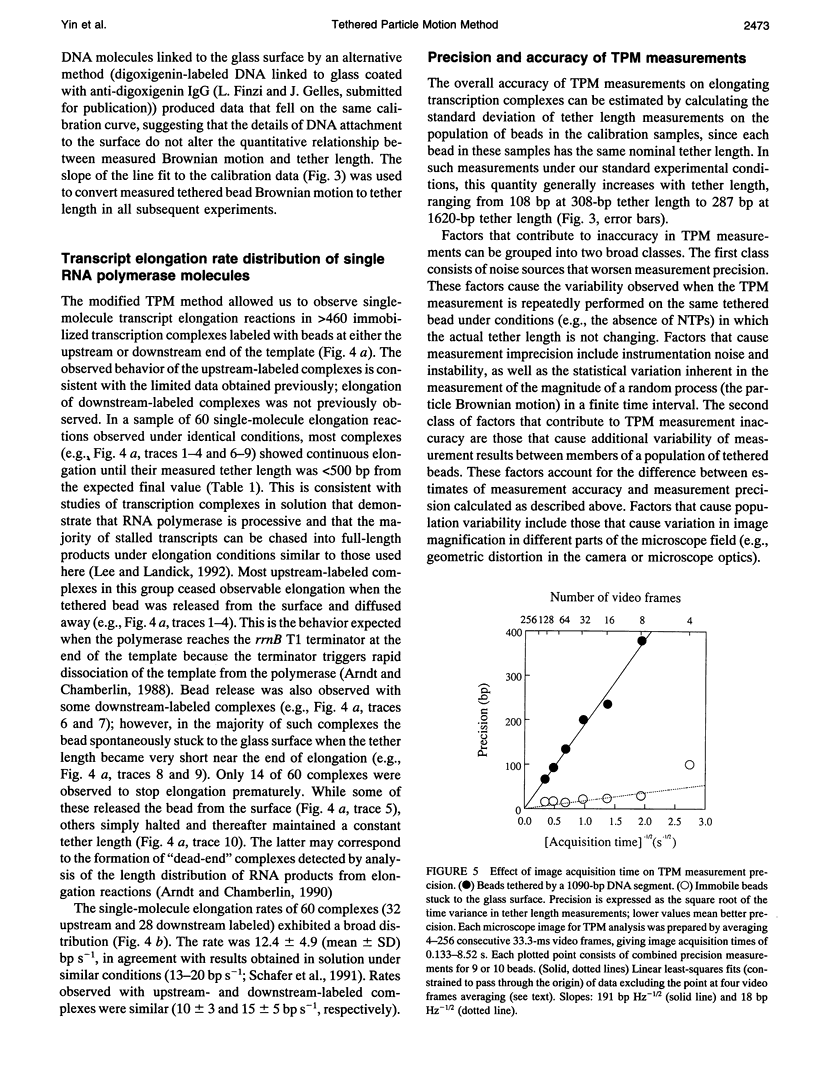
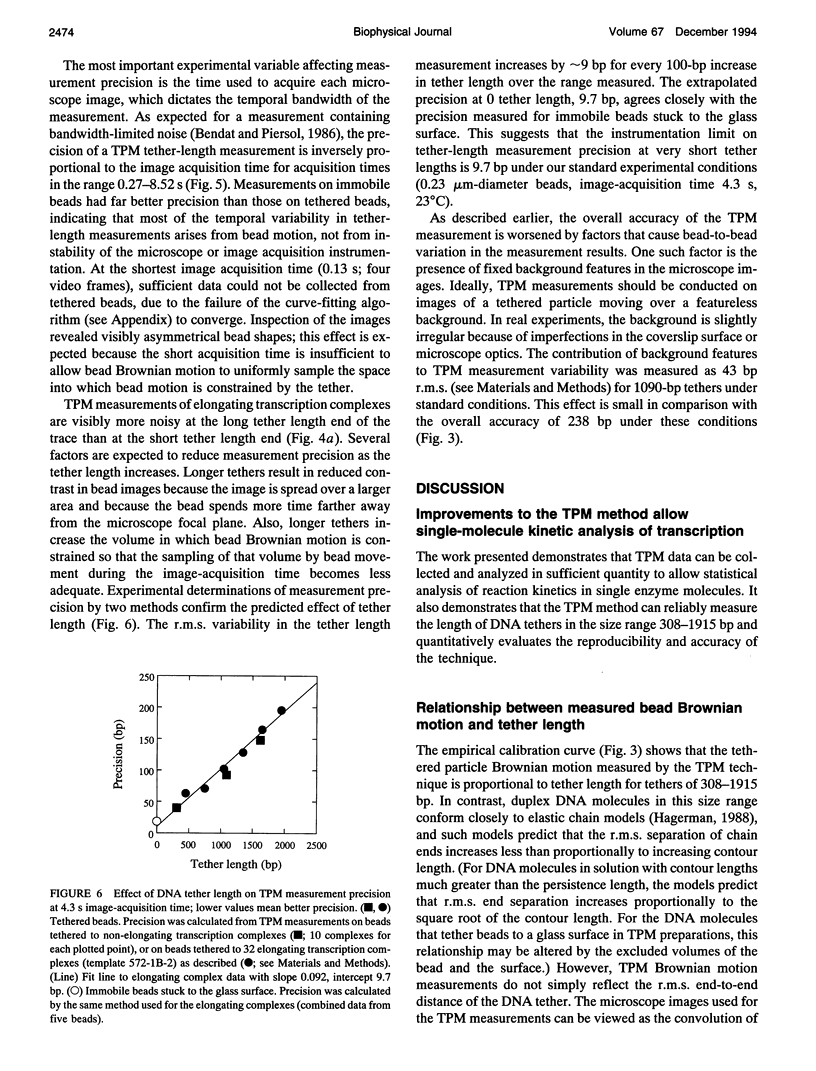
Schafer et al. (Nature 352:444-448 (1991)) devised the tethered particle motion (TPM) method to detect directly the movement of single, isolated molecules of a processive nucleic acid polymerase along a template DNA molecule. In TPM studies, the polymerase molecule is immobilized on a glass surface, and a particle (e.g., a 0.23 microns diameter polystyrene bead) is attached to one end of the enzyme-bound DNA molecule. Time-resolved measurements of the DNA contour length between the particle and the immobilized enzyme (the "tether length") are made by determining the magnitude of the Brownian motion of the DNA-tethered particle using light microscopy and digital image processing. We report here improved sample preparation methods that permit TPM data collection on transcript elongation by the Escherichia coli RNA polymerase at rates (approximately 10(2)-fold higher than those previously obtained) sufficient for practical use of microscopic kinetics techniques to analyze polymerase reaction mechanisms. In earlier TPM experiments, calculation of tether length from the observed Brownian motion was based on an untested numerical simulation of tethered bead Brownian motion. Using the improved methods, we have now empirically validated the TPM technique for tether lengths of 308-1915 base pairs (bp) using calibration specimens containing particles tethered by individual DNA molecules of known lengths. TPM analysis of such specimens yielded a linear calibration curve relating observed Brownian motion to tether length and allowed determination of the accuracy of the technique and measurement of how temporal bandwidth, tether length, and other experimental variables affect measurement precision. Under a standard set of experimental conditions (0.23 microns diameter bead, 0.23 Hz bandwidth, 23 degrees), accuracy is 108 and 258 bp r.m.s. at tether lengths of 308 and 1915 bp, respectively. Precision improves linearly with decreasing tether length to an extrapolated instrumentation limit of 10 bp r.m.s. and improves proportionally to the inverse square root of measurement bandwidth (1.9 x 10(2) bp Hz-1/2 for 1090-bp tethers). Measurements on large numbers of individual polymerase molecules reveal that time-averaged single-molecule elongation rates are more variable than is predicted from the random error in TPM measurements, demonstrating that the surface-immobilized RNA polymerase molecules are kinetically heterogeneous.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. M., Chamberlin M. J. RNA chain elongation by Escherichia coli RNA polymerase. Factors affecting the stability of elongating ternary complexes. J Mol Biol. 1990 May 5;213(1):79–108. doi: 10.1016/S0022-2836(05)80123-8. [DOI] [PubMed] [Google Scholar]
- Arndt K. M., Chamberlin M. J. Transcription termination in Escherichia coli. Measurement of the rate of enzyme release from Rho-independent terminators. J Mol Biol. 1988 Jul 20;202(2):271–285. doi: 10.1016/0022-2836(88)90457-3. [DOI] [PubMed] [Google Scholar]
- Berliner E., Mahtani H. K., Karki S., Chu L. F., Cronan J. E., Jr, Gelles J. Microtubule movement by a biotinated kinesin bound to streptavidin-coated surface. J Biol Chem. 1994 Mar 18;269(11):8610–8615. [PubMed] [Google Scholar]
- Chan C. L., Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989 Dec 5;264(34):20796–20804. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. The transcription termination site at the end of the early region of bacteriophage T7 DNA. Nucleic Acids Res. 1980 May 24;8(10):2119–2132. doi: 10.1093/nar/8.10.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erie D. A., Hajiseyedjavadi O., Young M. C., von Hippel P. H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993 Nov 5;262(5135):867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Erie D. A., Yager T. D., von Hippel P. H. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annu Rev Biophys Biomol Struct. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- FRAENKEL-CONRAT H., SNELL N. S., DUCAY E. D. Avidin. I. Isolation and characterization of the protein and nucleic acid. Arch Biochem Biophys. 1952 Jul;39(1):80–96. doi: 10.1016/0003-9861(52)90263-4. [DOI] [PubMed] [Google Scholar]
- Feng G. H., Lee D. N., Wang D., Chan C. L., Landick R. GreA-induced transcript cleavage in transcription complexes containing Escherichia coli RNA polymerase is controlled by multiple factors, including nascent transcript location and structure. J Biol Chem. 1994 Sep 2;269(35):22282–22294. [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J Mol Biol. 1984 Jan 25;172(3):263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- Garlick R. K., Giese R. W. Avidin binding of radiolabeled biotin derivatives. J Biol Chem. 1988 Jan 5;263(1):210–215. [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Titus G., Montibeller J. A., Finn F. M. Avidin binding of carboxyl-substituted biotin and analogues. Biochemistry. 1982 Mar 2;21(5):978–984. doi: 10.1021/bi00534a024. [DOI] [PubMed] [Google Scholar]
- Hsieh W. T., Whitson P. A., Matthews K. S., Wells R. D. Influence of sequence and distance between two operators on interaction with the lac repressor. J Biol Chem. 1987 Oct 25;262(30):14583–14591. [PubMed] [Google Scholar]
- Jencks W. P. The utilization of binding energy in coupled vectorial processes. Adv Enzymol Relat Areas Mol Biol. 1980;51:75–106. doi: 10.1002/9780470122969.ch2. [DOI] [PubMed] [Google Scholar]
- Jin D. J., Gross C. A. RpoB8, a rifampicin-resistant termination-proficient RNA polymerase, has an increased Km for purine nucleotides during transcription elongation. J Biol Chem. 1991 Aug 5;266(22):14478–14485. [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. RNA polymerase: regulation of transcript elongation and termination. FASEB J. 1991 Oct;5(13):2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992 May 20;225(2):239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Individual complexes halted along different transcription units have distinct and unexpected biochemical properties. J Mol Biol. 1992 May 20;225(2):221–237. doi: 10.1016/0022-2836(92)90917-9. [DOI] [PubMed] [Google Scholar]
- Landick R., Stewart J., Lee D. N. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990 Sep;4(9):1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- Lee D. N., Landick R. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J Mol Biol. 1992 Dec 5;228(3):759–777. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Chamberlin M. J. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987 Jul 5;196(1):61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Kassavetis G. A., Geiduschek E. P. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994 Jan 28;235(4):1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- Morris R. E., Saelinger C. B. Problems in the production and use of 5 nm avidin-gold colloids. J Microsc. 1986 Aug;143(Pt 2):171–176. doi: 10.1111/j.1365-2818.1986.tb02775.x. [DOI] [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Rhodes G., Chamberlin M. J. Ribonucleic acid chain elongation by Escherichia coli ribonucleic acid polymerase. I. Isolation of ternary complexes and the kinetics of elongation. J Biol Chem. 1974 Oct 25;249(20):6675–6683. [PubMed] [Google Scholar]
- Schafer D. A., Gelles J., Sheetz M. P., Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991 Aug 1;352(6334):444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. Amplification and isolation of Escherichia coli nusA protein and studies of its effects on in vitro RNA chain elongation. Biochemistry. 1984 Jan 17;23(2):197–203. doi: 10.1021/bi00297a004. [DOI] [PubMed] [Google Scholar]


