Abstract
Telomeres are usually maintained about an equilibrium length, and the set point for this equilibrium differs between species and between strains of a given species. To examine the requirement for telomerase in mediating establishment of a new telomere length equilibrium, we generated interspecies crosses with telomerase mTR knockout mice. In crosses between C57BL/6J (B6) and either of two unrelated mouse species, CAST/Ei and SPRET/Ei, telomerase mediated establishment of a new telomere length equilibrium in wild-type mTR+/+ mice. This new equilibrium was characterized by elongation of the short telomeres of CAST/Ei or SPRET/Ei origin. In contrast, mTR−/− offspring of interspecies crosses failed to elongate telomeres. Unexpectedly, haploinsufficiency was observed in mTR+/− heterozygous interspecies mice, which had an impaired ability to elongate short SPRET/Ei or CAST/Ei telomeres to the new equilibrium set point that was achieved in wild-type mTR+/+ mice. These results demonstrate that elongation of telomeres to a new telomere set point requires telomerase and indicate that telomerase RNA may be limiting in vivo.
Telomeres are DNA–protein complexes at the ends of linear eukaryotic chromosomes that play a critical role in maintaining chromosome stability. In mammalian cells, telomere DNA sequences consist of hundreds of tandem copies of the hexanucleotide repeat (TTAGGG)n. The average telomere restriction fragment length ranges among species from 10 to 15 kb in human to >50 kb in some Mus musculus strains. In the absence of compensatory mechanisms, telomeres shorten with each cell division because of incomplete replication of lagging strand DNA (1, 2). Human cells, when their telomeres become critically short, lose the capacity to divide and enter into cellular senescence (3).
Telomerase is a unique RNA-dependent DNA polymerase that synthesizes telomeric sequence repeats (4). Telomere elongation by telomerase compensates for telomere loss that occurs with cell division. Thus, telomere length is maintained as an equilibrium between processes that shorten and those that lengthen telomeres. This equilibrium is maintained around a specific “set point” that is species, and often strain, specific (1, 2). Regulation of this set point involves a number of different genes (5–8). Telomere binding proteins play a key role in regulating the set point, and alteration in the amount of telomere protein bound to telomeres can alter the set point (9–11). In the absence of telomerase, telomere length can no longer be maintained at the established set point.
Telomerase contains two essential components, the catalytic protein component (telomerase reverse transcriptase or TERT) and an RNA component (telomerase RNA or TR) that provides the template for telomere synthesis (1, 2). The physiologic role for telomerase has been demonstrated by studies in which one of the telomerase components is genetically inactivated and the consequences analyzed. Mice in which the RNA component mTR is deleted lack telomerase activity and initially show no detectable phenotypic change (12). However, after multiple generations of breeding, homozygous mTR−/− mice have significantly shortened telomeres and show increased frequency of chromosomal fusions (13). Apoptosis is the predominant cellular consequence of short telomeres in these mice. Germ cell apoptosis is seen in fourth generation mTR-deficient (G4) mice and increases in severity up to the sixth generation, which is functionally infertile (13, 14). Increased apoptosis is also seen in in vitro activated leukocytes from late generation mice (13).
Crosses between the mouse species Mus musculus domesticus, which has long telomeres (≈50 kb) and SPRET/Ei, which has shorter telomeres (10–15 kb), result in significant elongation of the short SPRET/Ei telomeres (7). Segregation analysis showed that a recessive gene on distal chromosome 2, unlinked to genes encoding known telomerase or telomere-associated components, plays a role in telomere elongation. These findings demonstrated that telomere lengthening can occur during normal development.
To analyze the mechanism mediating telomere elongation, we examined whether telomerase is required for elongation of short telomeres in crosses between M. musculus domesticus strain C57 BL/6 (B6) and either of two short-telomere mice, Mus Spretus (SPRET/Ei) or Mus musculus castaneus (CAST/Ei). We found that the increases in telomere length that occur in these crosses are mediated by telomerase. Unexpectedly, heterozygous mTR+/− mice were impaired in the ability to increase telomere length, suggesting that limiting telomerase may result in haploinsufficiency. Recent evidence suggests that hTR haploinsufficiency may be a cause of the human disease autosomal dyskeratosis congenita (15). Our results support the conclusion that a deficiency in telomerase RNA can lead to a deficiency in telomere elongation.
Methods
Mice.
M. musculus domesticus C57BL/6(B6), M. spretus (SPRET/Ei), and M. Musculus castaneus (CAST/Ei) mice were obtained from The Jackson Laboratory. These strains and crosses were bred at Bioqual (Rockville, MD) and at The Johns Hopkins School of Medicine (Baltimore). mTR-deficient mice were derived as previously described (12) and maintained as mTR heterozygotes on a B6 background (B6.mTR). mTR genotypes (wild type, +/+; heterozygous, +/−; or deficient, −/−) were identified with tail DNA purified by using a Dneasy tissue kit (Qiagen, Chatsworth, CA) and a three-primer PCR system. PCR reactions were run on a GeneAmp PCR machine (Perkin–Elmer) by using the following cycling conditions: 95°C, 10 min; 94°C, 1 min; 63°C, 1 min; 72°C, 1 min (40×); and 72°C, 10 min. Reactions were carried out in PCR buffer containing 1.5 mM MgCl2 (Perkin–Elmer), 0.2 mM dNTPs, 0.5 unit of Taq Gold polymerase (Perkin–Elmer), 2 μl of purified tail DNA, and three primers [wild-type forward (5′-agtgtctcggtgccttgact-3′), 1 μM; knock-out forward (5′-taccggtggatgtggaatgt-3′), 0.25 μM; and reverse (5′-gtgatgttgagttcccacag-3′), 1 μM]. Wild-type genomic mTR DNA is identifiable as a 367-nt band and KO mTR DNA as a 196-nt band; mTR heterozygous DNA yields both bands. Experiments were performed by using 3–8-month-old mice.
Pulsed-Field Gel.
DNA plugs were made from liver or spleen cells, enzyme digested, and subjected to pulsed- field gel electrophoresis as previously reported (7, 16).
In-Gel Hybridization.
Following electrophoresis and ethidium bromide staining, gels were dried on filter paper for 1 h at 50°C. Gels were prehybridized for 1 h at 55°C in 20 mM NaH2PO4/0.1% SDS/5 × Denhardt's Reagent/5 × SSC and hybridized with probe for 3 h at 55°C in 5 ml of prehybridization solution. (CCCTAA)4 oligonucleotides were obtained from Operon Technologies (Alameda, CA). Oligonucleotides were end-labeled with [γ-32P]ATP and then purified by using NAP-5 columns (Pharmacia Biotech). After hybridization, gels were washed three times for 20 min in 3 × SSC at room temperature and three times for 20 min in 3 × SSC/0.1% SDS at 58°C. Images were acquired with a Fuji Film BAS 1500 bioimaging analysis system with IMAGE READER software (version 1.3E).
Quantitative Fluorescence in Situ Hybridization (Q-FISH).
Spleen cells were stimulated in vitro for 48 h with Con A, lipopolysaccharide, and rIL2; 10 μg/ml Colcimide was added during the last 30 min of culture. Arrested cells were centrifuged for 10 min at 300 × g, resuspended in 0.4% KCl, and swelled for 4 min at 37°C. After hypotonic treatment, an equal volume of 3:1 methanol:acetic acid was added to each tube. Cells were then pelleted and washed in fresh fixative at least four times before by using. Cells were dropped onto clean glass slides to generate metaphase spreads. PNA FISH was performed with a Cy3-labeled (CCCTAA)3 PNA oligonucleotide (PE Biosystems) as described (17), and chromosomes were counterstained with DAPI (Sigma). FluoreSphere fluorescent beads (Molecular Probes) were used to monitor signal intensity loss during microscope use. Signal intensities were calculated by using designated Q-FISH software provided by the Lansdorp laboratory (17). Telomere signal intensities were measured on at least 10 metaphases for each mouse.
Telomerase Activity.
Telomerase was measured with an Invitrogen kit and a nonradioactive PCR-based telomeric repeat amplification protocol (TRAP) modified from the procedure originally described by Kim et al. (18). In vitro activation conditions for induction of telomerase activity in lymphocytes have been described previously (19). Briefly, single cell suspensions of spleen mononuclear cells were isolated from age-matched (B6.mTR+/− × SPRET/Ei) × B6.mTR+/− mice that were genotypically mTR+/+, mTR+/−, or mTR−/−. Three × 106 cells/ml were activated with a mixture of CD3-specific mAb (2C11) (5 μg/ml), lipopolysaccharide (10 μg/ml), and IL-2 (20 units/ml). After 60 h of culture, cells were harvested, counted, and lysed in Chaps buffer. Cell lysates were serially diluted and examined for telomerase activity. A portion of each lysate was heated to 85°C to inactivate telomerase and was included as a control in each assay. Reaction products were resolved on 10% acrylamide gels, stained with SyberGreen (Molecular Probes), and detected with a PhosphorImager (Molecular Dynamics).
RNA Isolation.
RNA was isolated from testes of individual 3–6-month-old mice by using an Rneasy kit (Qiagen) for (B6 × SPRET/Ei) crosses or with TriZol reagent (Life Technologies, Grand Island, NY) for (B6 × CAST/Ei) crosses. After isolation, RNA was treated with Rnase-free DNase (Promega), phenol∷chloroform extracted, precipitated, and resuspended in 10 mM Tris/0.5 mM EDTA (pH 8). RNA concentrations were determined by OD260 with a GeneSpec1 spectrophotometer (Hitachi, Tokyo). RNA integrity was confirmed by running a small RNA sample from each mouse on a 2% agarose gel, and RNA was visualized with ethidium bromide.
Real-Time PCR.
Real-time PCR was performed with the Taqman Gold kit (Applied Biosystems) and a two-step reaction. Primers used for reverse transcription and PCR include mTR-24F 5′-tgtgggttctggtcttttgttctccg-3′, mTR-134R 5′-gtttttgaggctcgggaacgcg-3′, mTR-24F (SPRET) 5′-tgtgggttctggtcttttgttcttcg-3′, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) F 5′-ttcaccaccatggagaaggc-3′, and GAPDH R 5′-ggcatggactgtggtcatga-3′ (20). Taqman probes were synthesized by Biosearch Technologies and included mTR 51T 5′-FAM-ccgctgtttttctcgctgacttccagcg-BHQ-1-3′ and mGAPDH 5′-ROX-tgcatcctgcaccaccaactgcttag-BHQ-2-3′.
To synthesize cDNA, 50 ng to 2.5 μg of total RNA was mixed with 300 nM each of mTR-134R and GAPDH R in a 50-μl reaction, heated for 5 min at 80°C, and chilled on ice. A 20-μl RT reaction contained 10 μl of RNA/primer, 0.25 unit of multiscribe reverse transcriptase, 0.2 unit of Rnase inhibitor, 1 × RT buffer, 5.5 mM MgCl2, and 2 mM dNTPs. Reactions were incubated for 50 min at 48°C, RT heat-inactivated at 85°C for 5 min, and cDNA either used immediately for PCR or stored at −20°C. Initially, genomic DNA contamination was assessed for each sample by using a control in which no RT was added (−RT). All RNA samples were found not to have significant DNA contamination as fluorescent intensities for GAPDH and mTR in −RT controls were not above background levels.
Each PCR reaction contained 1 × Taq Gold buffer (no ROX dye)/5.5 mM MgCl2/1 mM dNTPS/600 nM each of mTR and GAPDH primers/200 nM mTR probe/1.2 μM GAPDH probe, 0.5 unit of Taq Gold polymerase/5 μl of cDNA. PCR reactions were run on a Smart Cycler (Cepheid) with the following cycling conditions: 95°C, 10 min; 95°C, 15 s; 55°C, 30 s; 72°C, 45 s (40×); 72°C, 3 min. PCR products were initially run on agarose gels to confirm equimolar ratios of mTR and GAPDH products. Cycle thresholds were determined by using the second derivative of each curve for each dye channel. GAPDH was used as a loading control to monitor RT efficiency and RNA amount. Only samples that had similar and reproducible GAPDH thresholds (±0.5 cycles) for each input RNA concentration were used for quantitative purposes.
For quantitation, SPRET/Ei and CAST/Ei wild-type RNA was diluted to 500, 250, 100, and 50 ng/RT reaction. cDNA generated from these reactions was used to create a standard curve by using the input RNA concentration from the RT reaction and the threshold cycle from PCR. Slopes from respective standard curves were used to determine RNA concentrations from mTR+/+ and mTR+/− crosses by using the cycle threshold. Each RT reaction was repeated at least two independent times, and PCR reactions were repeated a minimum of four times.
Telomerase RNA Sequencing.
PCR amplification of telomerase RNA sequences was performed with primers mTR-pF 5′-GTGGGAAGTGCACCCGGAACTCGGTTC-3′ and mTR-pR 5′-CCCAGAAGTCGCAGGTTCTAGCGGC-3′ for 35 cycles of 95°C, 15 s; 60°C, 1 min; 72°C, 1 min. PCR products from two B6, two SPRET/Ei, and two CAST/Ei mice were sequenced.
Results
Establishment of a New Telomere Set Point Occurs in Interspecies Crosses.
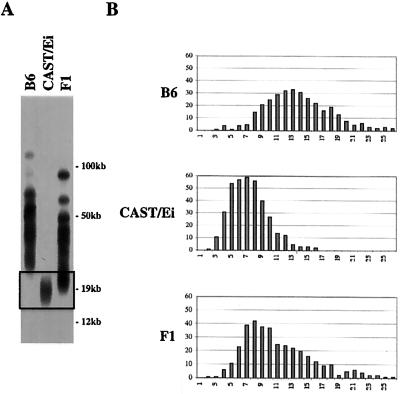
Some M. musculus mouse strains, such as B6, have characteristically long telomeres, with an average length of ≈50 kb. In contrast, SPRET/Ei and CAST/Ei have shorter telomere length distributions (7, 15, 21, 22). As previously demonstrated (B6 × SPRET/Ei), mice have a bimodal distribution of telomere length as assayed by pulsed field gel Southern analysis or Q-FISH (data not shown, ref. 7). The shorter set of telomeres, presumably of SPRET/Ei origin, is significantly longer in (B6 × SPRET/Ei) mice than are telomeres from parental SPRET/Ei mice, indicating that significant telomere elongation occurred during normal development. No detectable elongation of longer, presumably B6, telomeres was observed, suggesting that elongation is restricted to the shortest telomeres. To determine whether telomere elongation in interspecies crosses is specific to SPRET/Ei or reflects a more general mechanism of telomere length regulation, we crossed B6 and CAST/Ei mice. Southern analysis of (B6 × CAST/Ei) mice similarly demonstrated significant lengthening of the shorter CAST/Ei telomeres (Fig. 1A). The length of individual telomeres in parental and F1 mice was also assessed by Q-FISH, a technique that measures the amount of TTAGGG sequence in individual metaphase telomeres (17). In (B6 × CAST/Ei) mice, lengthening of the shorter telomeres was seen (Fig. 1B). Lengthening of shorter telomeres in interspecies crosses thus seems to be a general phenomenon associated with establishment of new telomere set points.
Figure 1.
CAST/Ei telomeres are elongated in offspring of a CAST/Ei × B6 cross. Telomere restriction fragments (TRF) from a B6, a CAST/Ei, and a (B6 × CAST/Ei) F1 mouse are shown. (A) End-labeled (CCCTAA)3 probe was used in in-gel hybridization to DpnII digests of agarose-embedded genomic DNA. The boxed region represents the shortest telomeres where differences in the distributions are most evident. (B) Q-FISH analysis of B6, CAST/Ei, and (B6 × CAST/Ei) F1 telomeres. Frequency distributions of 800 telomere signals from each mouse are shown.
Telomerase Is Required to Establish a New Length Equilibrium in Interspecies Crosses.
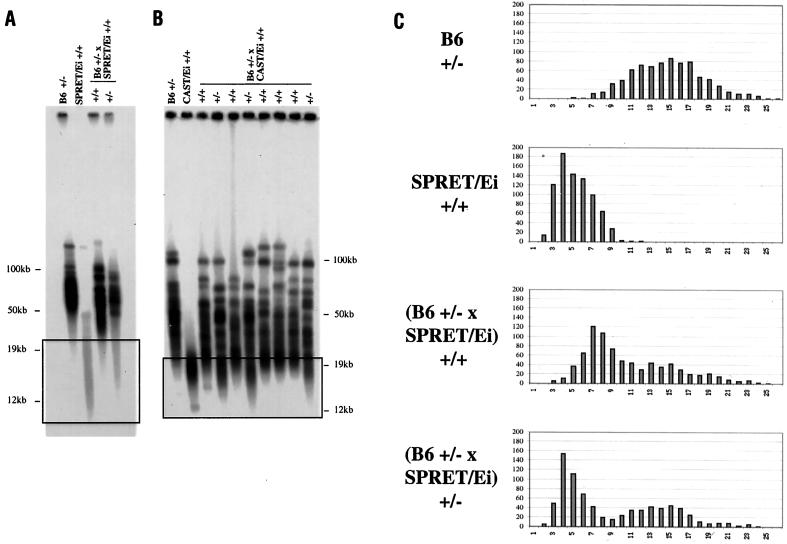
To determine whether telomerase is required for telomere elongation in interspecies crosses, we crossed both SPRET/Ei and CAST/Ei to heterozygous mTR-deficient (mTR+/−) B6 mice. Telomere length analysis of (B6 × SPRET/Ei) F1 mice showed that in mTR+/+ F1 mice, the short telomeres were elongated as had been seen previously (Fig. 2 A and C). It was anticipated that mTR heterozygous +/− mice would have intact telomerase activity and would therefore express wild-type telomere elongation. Unexpectedly, however, heterozygous mTR+/− (B6 × SPRET/Ei) F1 mice exhibited an impaired ability to increase the length of SPRET/Ei telomeres (Fig. 2 A and C).
Figure 2.
Elongation of short telomeres is impaired in mTR+/− offspring of B6 mTR+/− × SPRET/Ei and B6 mTR+/− × CAST/Ei crosses. (A) TRF distributions from the B6 mTR+/− and SPRET/Ei parents and mTR+/+ and mTR+/− offspring of a B6 mTR+/− × SPRET/Ei cross. End-labeled (CCCTAA)3 probe was used in in-gel hybridization to DpnII digests of agarose-embedded genomic DNA. The boxed region represents the shortest telomeres where differences in the distributions are most evident. (B) TRF distributions from the B6 mTR+/− and CAST/Ei parents and mTR+/+ and mTR+/− offspring of a B6 mTR+/− × CAST/Ei cross. (C) Q-FISH analysis of B6 mTR+/−, SPRET/Ei, and (B6 × SPRET/Ei) mTR+/+ and mTR+/− telomeres. Frequency distributions of 1,600 telomere signals from two mice of each genotype are shown.
We also crossed CAST/Ei with mTR+/− B6 mice. Whereas all mTR+/+ offspring showed substantial elongation of the short telomere class, mTR+/− offspring were deficient in elongation of these telomeres (Fig. 2B). This outcome was consistent independent of the male or female parental origin of the intact mTR allele in these crosses. These results indicate that telomere elongation in these crosses is mediated by telomerase. In addition, the lack of elongation to the new set point in mTR+/− mice suggests that telomerase RNA may be limiting in mTR+/− heterozygotes.
Telomere Shortening Is Seen in Telomerase Null Offspring from Interspecies Crosses.
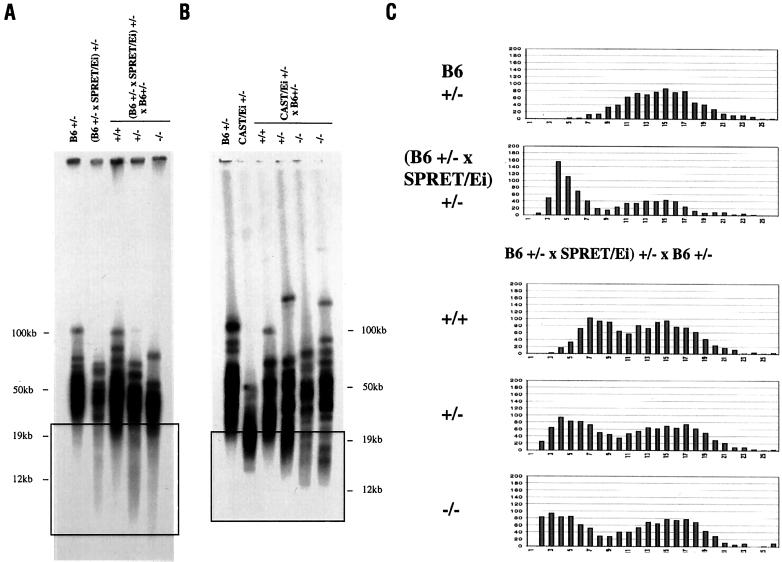
To further study the relationship between telomerase activity and telomere length, interspecies heterozygous mTR+/− (B6 × SPRET/Ei) and mTR+/− (B6 × CAST/Ei) mice were crossed with mTR+/− B6 mice, generating animals that were mTR wild-type (+/+), heterozygous (+/−), or homozygous-deficient (−/−). In mTR wild-type (+/+) [(B6 × SPRET/Ei) × B6] mice, a distribution of telomere lengths comparable to that seen in mTR wild-type (B6 × SPRET/Ei) mice was seen by pulsed field gel analysis (Fig. 3A). In mTR heterozygous (+/−) [(B6 × SPRET/Ei) × B6] mice, the shortest telomeres were maintained at a length similar to that observed in parental SPRET/Ei. Thus, there was a deficiency in elongation of these telomeres that was similar to the deficiency seen in the initial mTR+/− (B6 × SPRET/Ei) mice. Telomere length profiles in homozygous mTR-deficient (−/−) [(B6 × SPRET/Ei) × B6] mice showed telomere shortening such that the shortest telomeres in these mice were shorter than those of the SPRET/Ei parent, as seen in the boxed area of Fig. 3A. Q-FISH analysis of telomeres from these mice showed a bimodal distribution of telomere lengths (Fig. 3C). mTR+/+ [(B6 × SPRET/Ei) × B6] mice had no telomeres in the shortest telomere class of 1–2 Telomere fluorescence units, whereas mTR+/− mice had more telomeres in this short range. Finally, mTR−/− [(B6 × SPRET/Ei) × B6] mice showed a substantial number of telomeres in the shortest telomere class (Fig. 3C).
Figure 3.
Telomeres shorten in mTR−/− offspring of crosses between B6 mTR+/− and mTR+/− mice with short telomeres. (A) TRF distributions from B6 mTR+/− and (B6 mTR+/− × SPRET/Ei) mTR+/− parents and mTR+/+, mTR+/−, and mTR−/− offspring of a (B6 mTR+/− × SPRET/Ei) mTR+/− × B6 mTR+/− cross. End-labeled (CCCTAA)3 probe was used in in-gel hybridization to DpnII digests of agarose-embedded genomic DNA. The boxed region represents the shortest telomeres where differences in the distributions are most evident. (B) TRF distributions from B6 mTR+/− and CAST/Ei mTR+/− parents and mTR+/+, mTR+/−, and mTR−/− offspring of a B6 mTR+/− × CAST/Ei mTR+/− cross. CAST/Ei mTR+/− mice used here were bred by backcrossing mTR+/− (B6 × CAST/Ei) F1 mice three times to CAST/Ei. The boxed region represents the shortest telomeres where difference in the distributions are most evident. (C) Q-FISH analysis of telomeres from mTR+/+, mTR+/−, and mTR−/− offspring of a [B6 mTR+/− × (B6 mTR+/− × SPRET/Ei) mTR+/−] cross. Frequency distributions of 2,400 telomere signals from three mice of each genotype are shown.
Analogous experiments were carried out with CAST/Ei mice. mTR+/− (B6 × CAST/Ei) F1 mice were crossed to B6 mTR+/− mice. Again, the smallest visible telomeres were shorter in mTR−/− offspring than in mTR+/− littermates (Fig. 3B). The fact that telomeres in mTR−/− interspecies mice were shorter than telomeres from mTR+/− littermates suggests that some addition of telomere repeats occurs in mTR+/− animals. In addition, no chromosome fusions were seen in either mTR+/− or mTR−/− mice (data not shown), suggesting that telomere function was not compromised by limiting telomerase. Thus, regulation of telomere length represented by elongation to the new (B6 × SPRET/Ei) or (B6 × CAST/Ei) set point can be separated from maintenance of minimal telomere function.
mTR+/− Heterozygotes Have Reduced Telomerase RNA.
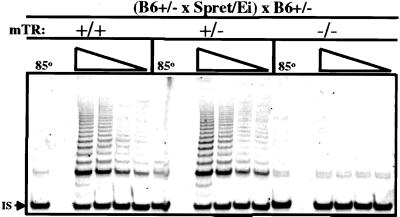
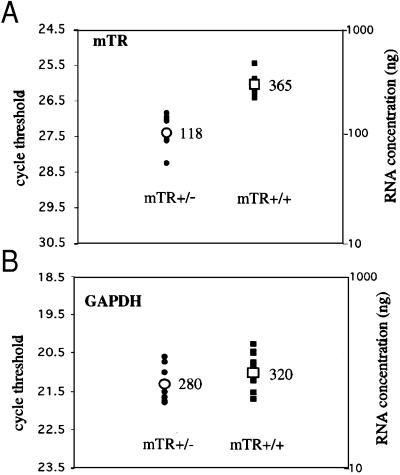
To determine whether the impaired lengthening of telomeres in mTR+/− mice was because of reduced telomerase activity, enzymatic activity was measured by in vitro TRAP assay. Telomerase activity was detected in lysates of activated splenocytes from [(B6 mTR+/− × SPRET/Ei) × B6mTR+/−] mice that were mTR+/+ or mTR+/−, but was undetectable in mTR−/− cells (Fig. 4). The TRAP assay showed no difference between telomerase activity levels in mTR+/+ and mTR+/− cells. However, this assay may not be sufficiently quantitative to detect small differences in telomerase activity. Thus, to examine whether telomerase RNA might be limiting in mTR+/− mice, we measured mTR levels by using quantitative real-time PCR. Total testis RNA from wild-type CAST/Ei was used to generate a standard curve. RNA was then isolated from three independent (B6 × CAST/Ei) mTR+/+ and mTR+/− littermates, and similar amounts of RNA were subjected to quantitative real-time PCR analysis. To control for the amount of RNA in each sample, a portion of the mouse GAPDH gene was amplified and showed similar PCR signal in all samples tested (Fig. 5). The level of RNA in mTR+/− samples was reproducibly less than half of that from mTR+/+ littermates (Fig. 5). Similar experiments were carried out with (B6 × SPRET/Ei) mTR+/+ and mTR+/− littermates, although the primers differed because of sequence variation in SPRET/Ei mTR (see below). The amount of mTR RNA in mTR+/− cells (81 ng) was again less than half that of mTR+/+ mice (260 ng). Thus, the lower level of telomerase RNA in mTR+/− heterozygotes may lead to the observed deficiency in telomere elongation in vivo.
Figure 4.
Similar levels of telomerase activity are expressed in mTR+/+ and mTR+/− mice. Lysates were prepared from in vitro activated spleen cells derived from age-matched [(B6 mTR+/− × SPRET/Ei) × B6 mTR+/−] mice that were genotypically mTR+/+, mTR+/−, or mTR−/− and serially diluted before analyzing telomerase activity. Telomerase activity is shown for 4,000, 2,000, 1,000, and 500 cell equivalents. As a control for telomerase activity, a sample (4,000 cell equivalents) was heated to 85°C for 10 min. IS, the band produced by the internal standard included in the TRAP reaction.
Figure 5.
mTR RNA levels are reduced in mTR heterozygous mice. Relative RNA concentrations (ng) were calculated from a standard curve generated from wild-type CAST/Ei mice (see Materials and Methods) and are indicated for each genotype. mTR levels were 365 in mTR+/+ and 118 in mTR+/− mice, and GAPDH levels were 320 in mTR+/+ and 280 in mTR+/− mice. Closed circles represent 10 independent threshold cycle values from mTR+/− mice, and the open circle represents the average. Closed squares represent 12 independent threshold cycle values from mTR+/+ mice, and the open square represents the average. (A) mTR. (B) GAPDH.
Species-Specific Differences in Telomerase RNA Sequence Are Not Responsible for Loss of Telomere Elongation.
In mTR+/− heterozygous (B6 × SPRET/Ei) F1 or (B6 × CAST/Ei) F1 mice, the single intact mTR gene is of SPRET/Ei or CAST/Ei origin, respectively. To examine whether potential sequence differences in mTR led to the deficiency in telomere lengthening, we sequenced mTR from B6, SPRET/Ei, and CAST/Ei. B6 and CAST/Ei mTR sequences were identical, whereas six sequence differences exist in the SPRET/Ei gene. These differences are C47/U, G80/A, G85/U, C154/U, C180/U, and C345/U. These changes are not in conserved regions of the telomerase RNA (23) and thus are not expected to lead to functional changes in telomerase activity. Because the RNA sequence was identical in B6 and CAST/Ei, the deficiency in telomere elongation observed in (B6 × CAST/Ei) F1 mice is not because of a defect in CAST/Ei mTR.
Additional data indicating that neither template-encoding nor noncoding (e.g., regulatory) differences between B6 and CAST/Ei mTR are responsible for loss of elongation came from observations of [(B6 × CAST/Ei) F1 × B6] animals. In mTR+/− mice from these crosses, the functional copy of mTR could derive from either parental species. If the species origin of mTR were causing impaired elongation, this would only be seen in 50% of the animals. However, a deficiency in telomere elongation was observed in each of 17 mTR+/− [(B6 mTR+/− × CAST/Ei) F1 × B6] mice. These results indicate that genetic polymorphisms linked to the mTR gene are not responsible for impaired telomere elongation observed in mTR+/− mice.
Discussion
A New Telomere Set Point Is Established in Interspecies Crosses.
In crosses between M. musculus mice with characteristically long telomeres (≈50 kb) and SPRET/Ei mice with shorter telomeres (10–15 kb), a dominant mechanism acts to elongate telomeres of SPRET/Ei origin in vivo (7). This mechanism establishes a new set point for telomere length in interspecies F1 animals. In the present study, we extended the analysis of interspecies telomere elongation. Crosses of B6 × CAST/Ei showed a similar elongation of the short CAST/Ei telomeres in F1 animals. Selective lengthening of short telomeres in interspecies crosses thus seems to be a general phenomenon.
Telomerase RNA Is Limiting in the Establishment of a New Set Point.
To characterize the role of telomerase in elongation of telomeres, we generated interspecies crosses in mice deficient for telomerase. Unexpectedly, we found a deficiency in SPRET/Ei and CAST/Ei telomere elongation to the new interspecies set point in mTR+/− heterozygotes. These results indicate that telomerase is required for elongation to the new set point and that the dose of mTR gene has a significant functional effect on telomere elongation. Quantitative analysis of mTR levels showed that mTR RNA is reduced in mTR+/− mice relative to the level in wild-type mTR+/+ mice. There thus exists a dose-response relation between mTR expression and functional telomere length maintenance in vivo.
The factors that determine telomere elongation in vivo are not fully understood. Although both TR and TERT are essential for telomerase activity, it is not known whether telomerase RNA or the catalytic telomerase reverse transcriptase (TERT) is quantitatively limiting in vivo. A recent report suggested that mTERT is limiting in murine embryonic stem cells because embryonic stem cells that are heterozygous for mTERT deficiency undergo accelerated telomere shortening (24). The data presented here suggest that the RNA component may be limiting under certain in vivo conditions.
Telomere Function Is Maintained Even with Limiting Telomerase.
To compare the deficiency in elongation in mTR+/− interspecies mice to a situation in which there is complete loss of telomerase function, we backcrossed mTR+/− interspecies mice to B6 mTR+/− mice to generate [(B6 × SPRET/Ei) × B6] and [(B6 × CAST/Ei) × B6] mice with the genotypes mTR+/+, mTR+/−, and mTR−/−. mTR+/+ mice showed similar telomere elongation to that seen in mTR+/+ (B6 × SPRET/Ei) F1 mice. mTR+/− [(B6 × SPRET/Ei) × B6] and [(B6 × CAST/Ei) × B6] mice had short telomeres that were not different in length from telomeres present in parental SPRET/Ei and CAST/Ei mice, as had been observed in mTR+/− F1 mice. In contrast, mTR−/− mice showed substantial shortening of the short SPRET/Ei and CAST/Ei-derived telomeres. In all metaphases examined by Q-FISH, TTAGGG sequences were visible on all telomeres in both mTR+/− and mTR−/− mice. In addition, no chromosome fusions were seen in these mice, indicating that telomere function was not compromised by the limiting telomerase. It was notable that, even with limiting telomerase, there were fewer telomeres in the shortest class in mTR+/− mice from [(B6 × SPRET/Ei) × B6] and [(B6 × CAST/Ei) × B6] crosses compared with their mTR−/− littermates. This suggests that telomerase may be specifically recruited to the shortest telomeres even when the telomere set point cannot be maintained. These conclusions are similar to those described for heterozygous mTERT+/− embryonic stem cells. Although both mTERT−/− and mTERT+/− embryonic stem cells show telomere shortening, telomere signal is not lost from the ends of mTERT+/− chromosome, whereas it is lost from mTERT−/− chromosomes (25). Thus, the maintenance of a specific telomere length set point is separable from maintenance of telomere function.
Haploinsufficiency May Cause a Phenotype After a Lag.
The recent discovery that the autosomal dominant disease dyskeratosis congenita is due to deletion of one copy of the telomerase RNA gene (15) provides additional evidence for phenotypic consequence of telomerase RNA haploinsufficiency. Although we did not find evidence for loss of telomere function in the first or second generation of heterozygous mice, it is possible that a phenotypic lag in the appearance of telomere dysfunction may occur in these F1 heterozygous mice. This phenotypic lag would resemble that seen for mTR−/− mice (13), but it is likely to occur more slowly. Even though telomerase is targeted to the shortest telomeres, at a certain point the limiting telomerase may not be able to maintain the length of the large number of critically short telomeres. The onset of disease in dyskeratosis congenita patients was reported to occur at progressively younger ages in successive generations, suggesting that such a phenotypic lag in the appearance of consequences of haploinsufficiency may operate in this disease.
Acknowledgments
We thank Carl Barrett, Lea Harrington, Andre Nussenzweig, Michael Seldin, and Jennifer Hackett for critical reading of this manuscript. We thank Jiunn-liang Chen for advice on PCR cloning and real-time PCR of mTR and Genevieve Sanchez and staff at Bioqual for expert animal care and breeding. K.K.O. was supported by a Leukemia and Lymphoma Society Postdoctoral Fellowship. This work was sponsored in part by National Institutes of Health Grant CA16519 (to C.W.G.).
Abbreviations
- TERT
telomerase reverse transcriptase
- TR
telomerase RNA component
- TRF
telomere restriction fragment
- Q-FISH
quantitative fluorescence in situ hybridization
- TRAP
telomeric repeat amplification protocol
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
References
- 1.Greider C W. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 2.McEachern M J, Krauskopf A, Blackburn E H. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 3.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 4.Greider C W, Blackburn E H. Nature (London) 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 5.Walmsley R M, Petes T D. Proc Natl Acad Sci USA. 1985;82:506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burr B, Burr F A, Matz E C, Romero-Severson J. Plant Cell. 1992;4:953–960. doi: 10.1105/tpc.4.8.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Hathcock K S, Hande P, Lansdorp P M, Seldin M F, Hodes R J. Proc Natl Acad Sci USA. 1998;95:8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie K B, Mallory J C, Petes T D. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Steensel B, de Lange T. Nature (London) 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 10.Shore D. J Biochem (Tokyo) 1997;378:591–597. [Google Scholar]
- 11.Krauskopf A, Blackburn E H. Nature (London) 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 12.Blasco M A, Lee H-W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 13.Lee H-W, Blasco M A, Gottlieb G J, Horner J W, II, Greider C W, DePinho R A. Nature (London) 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 14.Hemann M T, Greider C W. Mol Biol Cell. 2001;12:2023–2030. doi: 10.1091/mbc.12.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason P J, Dokal I. Nature (London) 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 16.Hemann M T, Greider C W. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lansdorp P M, Verwoerd N P, van de Rijke F M, Dragowska V, Little M T, Dirks R W, Raap A K, Tanke H J. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 18.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 19.Hathcock K S, Weng N-p, Merica R, Jenkins M K, Hodes R J. J Immunol. 1998;160:5702–5706. [PubMed] [Google Scholar]
- 20.Overbergh L, Valckx D, Waer M, Mathieu C. Cytokine. 1998;11:305–312. doi: 10.1006/cyto.1998.0426. [DOI] [PubMed] [Google Scholar]
- 21.Kipling D, Cooke H J. Nature (London) 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 22.Starling J A, Maule J, Hastie N D, Allshire R C. Nucleic Acids Res. 1990;18:6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J L, Blasco M A, Greider C W. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Snow B E, Hande M P, Yeung D, Erdmann N J, Wakeham A, Itie A, Siderovski D P, Lansdorp P M, Robinson M O, Harrington L. Curr Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Kha H, Ungrin M, Robinson M O, Harrington L. Proc Natl Acad Sci USA. 2002;99:3597–3602. doi: 10.1073/pnas.062549199. [DOI] [PMC free article] [PubMed] [Google Scholar]