Abstract
Many Drosophila non-long terminal repeat (LTR) retrotransposons actively transpose into internal, gene-rich regions of chromosomes but do not transpose onto chromosome ends. HeT-A and TART are remarkable exceptions; they form telomeres of Drosophila by repeated transpositions onto the ends of chromosomes and never transpose to internal regions of chromosomes. Both telomeric and nontelomeric, non-LTR elements transpose by target-primed reverse transcription, and their targets are not determined simply by DNA sequence, so it is not clear why these two kinds of elements have nonoverlapping transposition patterns. To explore roles of retrotransposon-encoded proteins in transposition, we analyzed intracellular targeting of Gag proteins from five non-LTR retrotransposons, HeT-A, TART, jockey, Doc, and I factor. All were expressed as green fluorescent protein-tagged proteins in cultured Drosophila cells. These Gag proteins have high levels of sequence similarity, but they have dramatic differences in intracellular targeting. As expected, HeT-A and TART Gags are transported efficiently to nuclei, where they show specific patterns of localization. These patterns are cell cycle-dependent, disappearing during mitosis. In contrast, only a fraction of jockey Gag moves into nuclei, whereas neither Doc nor I factor Gag is detected in the nucleus. Gags of the nontelomeric retrotransposons form characteristic clusters in the cytoplasm. These experiments demonstrate that closely related retrotransposon Gag proteins can have different intracellular localizations, presumably because they interact differently with cellular components. We suggest that these interactions reflect mechanisms by which the cell influences the level of transposition of an element.
Most retrotransposons have two large ORFs with strong similarity to the gag and pol coding regions of retroviruses (Fig. 1). In both retrotransposons and retroviruses, the pol sequence encodes reverse transcriptase and other enzymatic activities necessary for transposition. Regions of the polypeptide responsible for each activity can be identified by conserved amino acid sequence motifs. In contrast, gag coding regions show less sequence similarity and functions of their products are less well understood. Extensive genetic and biochemical studies have shown that, despite sequence differences, Gag proteins from typical retroviruses have similar activities in packaging viral RNA and facilitating its export from the host cell. Parts of this protein, after proteolytic processing and perhaps other modification, are also essential for steps leading to integration of the virus in a new host; however, the details of Gag activities in infection are less well understood than its roles in virus production (reviewed in refs. 1 and 2). Gag proteins of many retrotransposons have striking similarities to retroviral Gags. Even less is known about roles of retrotransposon Gags than about activities of retroviral Gags during infection; however, the structural similarities suggest that there may be similarities in function.
Figure 1.
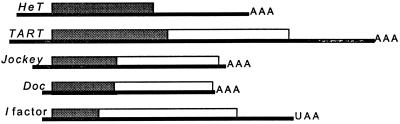
Diagrams of non-LTR retrotransposons in this study. Each element is represented as the RNA transposition intermediate, drawn approximately to scale. Thin lines indicate noncoding regions. Shaded boxes show gag coding regions. Open boxes are pol coding regions. HeT-A does not have a pol gene. AAA represents the poly(A) tail of the RNA.
The question of Gag protein function is especially interesting for the Drosophila telomeric retrotransposons HeT-A and TART because these elements have a defined role in chromosome structure yet have maintained characteristics of non-LTR (non-long terminal repeat) retrotransposons, including their gag and pol coding regions (reviewed in refs. 3 and 4). (HeT-A is an atypical element because it has no pol coding region, although it transposes actively.) HeT-A and TART form the telomeres by successive transpositions to the chromosome ends. They fulfill the role played by telomerase-generated repeats in other eukaryotes and, thus, are the first transposable elements with a defined role in chromosome structure. These elements are remarkable because they transpose only to ends of chromosomes. This contrasts sharply with other Drosophila non-LTR retrotransposons that have similar coding sequences but are never found in telomeres. Instead, these nontelomeric elements are active in transposing within euchromatic, gene-rich regions of the chromosomes. To determine whether the distinction between telomeric and nontelomeric elements is reflected in functions of their encoded proteins, we have compared Gag proteins from telomeric and nontelomeric retrotransposons.
Although initial ideas of the roles of Gag in retrotransposition came from analogy to retroviral proteins, there is now some evidence that retrotransposon Gags are involved in transposition. For two non-LTR retrotransposons, human LINE-1 and Drosophila I factor, it has been possible to achieve in vivo transposition of genetically modified elements (5, 6). Experiments with both systems have shown that Gag proteins are essential for transposition. Such experiments are not yet possible for the Drosophila telomere transposons. Nevertheless, sequence analyses on HeT-A elements suggest that the ability to translate Gag is necessary for successful transposition (7). The gag ORFs of HeT-A elements have a polymorphic region in which elements can differ by nucleotide changes and insertions/deletions. Remarkably, none of the changes in this region introduces a stop codon or a frameshift in any of the elements. Thus, despite the frequent sequence changes, at least in the polymorphic region, all elements that have transposed successfully have maintained the ability to produce an intact Gag protein, suggesting that the protein was involved in some step in the conversion of HeT-A RNA into a chromosomal DNA copy.
In the study reported here, we compare Gag proteins of five Drosophila non-LTR retrotransposons—the two telomeric elements and three of the best-known nontelomeric elements (Fig. 1). Two of the nontelomeric elements, Doc and jockey, are from the Jockey clade, the group that includes TART in the reverse transcriptase-based phylogeny of retroelements (8). HeT-A cannot be included in this classification because it does not encode reverse transcriptase; however, our studies of gag sequences show that HeT-A gag sequences are most similar to the sequences in this clade (7). Our third nontelomeric element, I factor, belongs to a distantly related clade. To make comparisons, we have expressed each of the proteins (Fig. 2) from the same promoter and in the same cell type. The constructs used differ only in their coding sequences, implicating the protein itself in the intracellular localization. Each of the proteins was fused to green fluorescent protein (GFP) so that it could be followed in cytological preparations.
Figure 2.
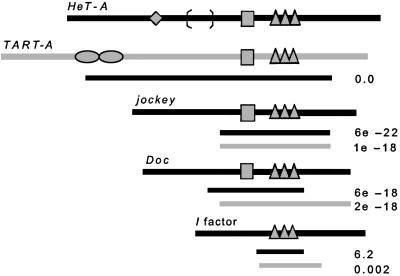
Diagrams of Gag proteins in this study, drawn approximately to scale, showing known motifs and landmarks. Striped boxes indicate the MHR region (not present in I factor). Three triangles indicate the three zinc knuckles present in each protein. Diamond on HeT-A and ovals on TART indicate length of polymorphic regions where proteins from different copies of the element differ in number of amino acids. Brackets on HeT-A mark limits of the segment of different amino acids in the 9D4 Gag. Black lines under each protein indicate region of amino acid sequence similarity between that protein and HeT-A Gag, defined by blast alignment. Gray lines under each protein mark blast alignment with TART Gag. Number at end of each line is the E value of the blast alignment.
We expected that, if Gags showed differences in localization, these would be in the nucleus, reflecting sites of transposition. This expectation was based partly on studies of the yeast LTR retrotransposon Ty5, which transposes into silent chromatin because of an interaction between its integrase and the chromatin protein, Sir4p (9). Gag proteins might have analogous roles in target site selection.
We did find element-specific patterns of localization within the nucleus for some Gags. Surprisingly, we also found element-specific patterns of localization in the cytoplasm. These different localizations suggest that Gags of different elements may not have the same interactions with cellular components as they travel from ribosome to final destination. The localizations we detect may reveal pathways of interactions between translation of the Gag protein and successful transposition of the element.
Methods
Recombinant DNA.
Each gag coding sequence was amplified by PCR with primers that included the initial Met codon placed within an optimal translation initiation sequence (CCACCATGT). The 3′ primers included the final codons fused in-frame with the GFP coding sequence. Each of the amplified coding sequences was inserted in vector pPL17, kindly provided by Ilaria Rebay (Whitehead Institute, Massachusetts Institute of Technology). pPL17 contains the EGFP coding sequence (CLONTECH) under the Drosophila armadillo promoter.
The HeT-A Gag ORF DNA was isolated from element 23Zn-1 (GenBank accession no. U06920). The variant HeT-A Gag ORF from element 9D4 (GenBank accession no. X68130) was isolated from a clone kindly provided by H. Biessmann (University of California, Irvine). The TART A ORF1 (Gag) initially was isolated from GenBank accession no. AY035776. This sequence contains a stop codon at nucleotide 208. The stop was replaced with a valine codon (GTA). Jockey ORF1, Doc ORF1, and I factor ORF1 coding sequences were isolated from clones, kindly provided by R. Levis (Carnegie Institution, Baltimore), I. Busseau (Institute de Genetique Humaine, Montpellier, France), and T. Heidmann (Institute Gustave-Roussy, Villejuif, France).
Transient Transfection.
Drosophila Schneider line 2 (SL2) cells were transfected with 5–10 mg of plasmid DNA, using a Cytofectene Transfection Reagent Kit (Bio-Rad). Transfection was in 2.5 ml of serum-free medium for 6 h. Cells were analyzed by fluorescence microscopy and immunoblotting 24 and 48 h after the start of transfection.
Immunoblot Analysis.
Protein samples from transfected cells were resolved by SDS/PAGE and transferred to poly(vinylidene difluoride) membranes (Bio-Rad). The fusion proteins were visualized by immunoblotting with anti-GFP antiserum and secondary antibody conjugated to alkaline phosphatase (Jackson ImmunoResearch). The polyclonal anti-GFP antiserum was from guinea pigs, immunized with purified GFP protein (CLONTECH). [Antibody was produced by Covance Research Products (Richmond, CA).]
Slide Preparation and Microscopy.
Cell suspensions were dropped on slides and fixed with 3.7% formaldehyde in PBT (PBS/0.1% Tween-20). Slides were washed with PBT, stained with 4′,6-diamidino-2-phenylindole (DAPI), and mounted in 50% glycerol/PBS. Localization of GFP-tagged proteins was analyzed with a Nikon ECLIPSE E 600 microscope equipped with a charge-coupled device camera. Fluorescent and differential interference contrast images were taken with SPOT RT 3.0 and processed with Adobe PHOTOSHOP 5.5 (Adobe Systems, Mountain View, CA).
Results
GFP-Tagged Gag Proteins from Drosophila Non-LTR Retrotransposons Are Expressed in Cultured Drosophila Cells.
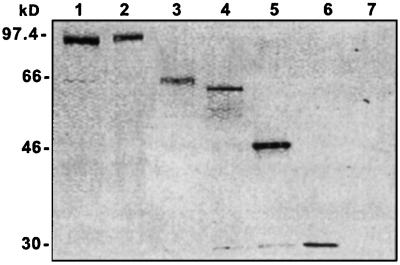
We have used transient transfection of cultured Drosophila cells (SL2 cells) to study Gags with C-terminal GFP tags. The C-terminal GFP tag should be detected only if the entire coding region is translated. Immunoblot analysis of protein from both 24- (Fig. 3) and 48-h samples showed that, for each construct, the only protein detected by an antibody against GFP was of the size expected for the full-length tagged protein. Gel lanes were loaded with equal numbers of cells and, with the exception of the jockey lane, appear to contain roughly equal amounts of GFP-tagged protein. The jockey protein was somewhat less abundant than the others. Because all proteins were expressed from the same promoter, we suppose that the lower amount of protein may reflect a lower stability of jockey Gag, rather than lower mRNA transcription. This supposition is supported by a further decrease in protein in the 48-h sample.
Figure 3.
Fusion proteins are stable and of the expected size. Total protein from transfected SL2 cells were separated by SDS/PAGE and analyzed by immunoblotting with anti-GFP antiserum to detect all GFP-tagged proteins. Expressed proteins were: 1, HeT-A Gag; 2, TART Gag; 3, jockey Gag; 4, Doc Gag; 5, I factor Gag; 6, GFP; 7, nontransfected cells. Molecular mass (kDa) is shown on the left.
The HeT-A Gag Protein Is Targeted to Defined Nuclear Domains and Forms a Specific Cytoplasmic Structure.
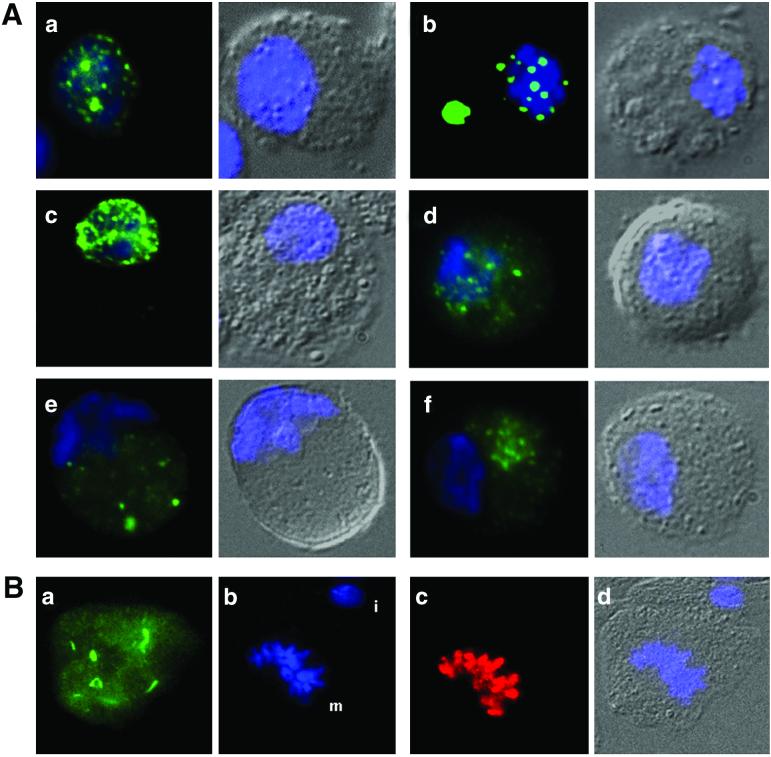
When the full-length HeT-A coding region of element 23Zn-1 is expressed in Drosophila cells, the protein is transported rapidly into the nucleus. Inside the nucleus, it forms clusters with a distinctive appearance. These are discrete clusters, relatively uniform in size and shape, which we refer to as “Het-dots.” We have detected nuclei that have either many small dots or fewer large ones. Some nuclei with small dots have a few large dots (Fig. 4A). The fraction of cells with large dots (Fig. 4B) increases with time after transfection, and we believe that this reflects the progressive formation of Het-dots, with the larger dots being the mature stage. Whether the dots are large or small, they are scattered through the nucleus, with a tendency to be near the nuclear membrane. Because telomeres tend to be located near the nuclear periphery, this localization pattern suggests that Het-dots may be at or near telomeres. In addition, in cells with large Het-dots, the number of dots is in the range expected for telomeres in these cells. Most cells in our SL2 line have a single X chromosome and three pairs of autosomes. They therefore would be expected to have no more than 14 telomere spots. In these small nuclei, fusion or superimposition of dots might occur frequently and decrease this number. Typically, 10–14 dots can be detected in each nucleus with large dots.
Figure 4.
Intracellular localization of GFP-tagged Gags in transiently transfected SL2 cells. (A) Fluorescence micrographs show each cell in two panels. DNA in all cells is stained with DAPI (blue). Left show merged GFP and DAPI. Right show differential interference contrast image with DAPI staining superimposed. Transfectants shown: a, HeT-A Gag—a cell with a large number of small clusters in the nucleus; b, HeT-A Gag—a cell with large Het-dots in the nucleus and Het-body in the cytoplasm; c, TART Gag—small clusters in the nucleus, so numerous that they do not resolve well in the picture; d, jockey Gag—irregular clusters in the nucleus and clusters plus diffuse protein in the cytoplasm; e, Doc Gag—irregular clusters in cytoplasm; f, I factor Gag—clusters in the cytoplasm. In d–f, the clusters are on a background of diffuse protein. For jockey Gag (D), this background does not extend completely to the edge of the cell. (B) Localization of HeT-A Gag at metaphase. Randomly cycling cells expressing HeT-A Gag were stained with an antibody against metaphase-specific phospho-H3 (red). Figure shows a cell in metaphase with part of an (nontransfected) interphase cell above it. The distinctive localizations of GFP-Gag seen in interphase have dispersed, and protein is spread throughout the cell, avoiding the condensed chromosomes. DNA is stained with DAPI. a, GFP; b, DAPI (m = metaphase chromosomes, i = interphase nucleus of adjoining cell); c, anti-H3-P (red) staining metaphase chromosomes; d, differential interference contrast image with DAPI.
In a fraction of the cells with Het-dots, we also see an unusual cytoplasmic accumulation of protein (Fig. 4B). Typically, this cytoplasmic Gag-GFP forms a single, oval-shaped structure, frequently with smooth edges, and much larger than the nuclear dots. This cytoplasmic structure, referred to as the “Het-body,” appears to be preferentially in cells with larger nuclear dots. The presence of the Het-body does not diminish the brightness of the nuclear Het-dots. It is detected either adjacent to or away from the nucleus. Because the body is seen only in cells with Het-dots, it appears to be a result of overexpression of the protein. If this is the case, formation of the Het-body in the cytoplasm may suggest that the nuclear targets of full-length HeT-A Gag are saturable.
The Het-dots and Het-body are specific for HeT-A Gag protein. These structures are also seen with a variant of the protein from another HeT-A element (9D4, see below), but are not detected with TART Gag nor any of the nontelomeric retrotransposon Gags we have studied.
A Variant HeT-A Gag with a Substitution Region of 55 aa Forms Het-Dots and Het-Bodies.
Early studies suggested that the HeT-A gag coding region might contain two ORFs, linked by a −1 frame shift (7, 10). However, we now know that this sequence is a single, continuous ORF. In the course of correcting this error, we have identified a HeT-A element with a variant ORF that has a 55- aa segment with almost no similarity to other HeT-A Gag proteins (3).
This variant ORF, in element 9D4 (11), has 94% nucleotide identity with the ORF of the 23Zn-1 element, used as our standard ORF. However, alignment of sequences shows a region in which an “extra” nucleotide in 9D4 changes the reading frame for 9D4 relative to 23Zn-1. The two sequences are read in different frames for 55 codons, until an “extra” nucleotide in 23Zn-1 moves the reading frames into synchrony. These two nucleotide differences result in two elements with a region of 55 codons having only 13% amino acid identity but 98% nucleotide identity (brackets in Fig. 2). The 9D4 protein essentially is a substitution mutation in comparison with the other HeT-A Gag proteins from both Drosophila melanogaster and Drosophila yakuba (7, 12). This region of 9D4 also has less similarity to the TART protein than does the 23Zn-1 sequence.
The large amino acid substitution in the 9D4 Gag has no detectable effect on the intracellular localization of the protein. When tagged with GFP, the full-length 9D4 protein localizes to Het-dots and forms Het-bodies indistinguishable from those of the 23Zn-1 protein (data not shown). The large sequence substitution also did not affect the ability of 9D4 to transpose. It was cloned shortly after it had moved onto the end of a healing broken chromosome (11). The 55 aa are in one of the more variable parts of the protein, a part that does not contain any of the conserved motifs shown in Fig. 2.
TART Gag Also Is Targeted to the Nucleus.
Like HeT-A Gag, TART Gag is moved rapidly into the nucleus. However, TART Gag has a qualitatively different distribution within the nucleus. It forms intranuclear clusters that are smaller and much greater in number than the Het-dots (Fig. 4C). These clusters are also similar in size in the nuclei of all transfected cells. A small amount of TART Gag is detected in cytoplasmic clusters but it does not form a structure like the Het-body.
Localization of jockey Gag to the Nucleus Is Slow and Inefficient.
Although HeT-A and TART Gags localized efficiently to nuclei, only a fraction of the jockey protein moved into the nucleus (Fig. 4D). Inside the nucleus, it is dispersed in small, irregular clusters, easily distinguishable from the structures formed by either HeT-A or TART. At later time points, the nuclear pattern of localization does not change, but much of the protein remaining in the cytoplasm forms large clusters, frequently two per cell (not shown). The protein in the cytoplasm is not uniformly dispersed. Instead, it seems to be less concentrated near the edges of the cell.
Gag Proteins from Doc and I Factor Do Not Appear to Enter the Nucleus.
Both Doc and I factor Gag proteins are dispersed throughout the cytoplasm and appear to be completely absent from the nucleus (Fig. 4 E and F). However, we cannot exclude the possibility that cytological techniques could easily miss small amounts of these proteins. In the cytoplasm, both proteins form clusters within a background of more diffuse protein. The I factor clusters are small and similar in size, whereas the Doc clusters are irregular in size and shape.
The Localization of HeT-A and TART Gags Is Dynamic and Changes in a Cell Cycle-Dependent Manner.
During metaphase, the localization of both HeT-A and TART Gags changes dramatically. The change is similar for the two proteins: the nuclear clusters as well as the Het-body are dispersed and the two proteins become diffused through the cell, except in the area of the condensed chromosomes (Fig. 4). Only random streaks of accumulated material are seen on the background of evenly spread protein. In cells in which the nuclear membrane has formed around the two new nuclei, HeT-A Gag is detected in small nuclear dots, presumably the initial steps in the formation of large Het-dots. It is unclear whether the material in the daughter nuclei is newly synthesized or recycled.
In contrast to the dramatic change in the localization of HeT-A and TART Gags, proteins of the nontelomeric retrotransposons did not undergo visible change. Instead, they remained in clusters within a background of diffuse protein, distributed throughout the cell.
To avoid disrupting cell morphology, our analyses of cell cycle-related changes were done on randomly cycling cells without colchicine or other drugs to trap cells in mitosis. Mitotic stages were identified on morphological criteria, aided by staining with an antibody that recognizes the metaphase-specific, phosphorylated histone H3.
Discussion
Closely Related Retrotransposon Gag Proteins Can Have Different Intracellular Targeting.
Retrotransposons generally are considered selfish, or parasitic, elements because they encode only proteins useful for their own transposition. The two Drosophila retrotransposons, HeT-A and TART, are the only clear exceptions to this classification. Our studies of Gag proteins now show that the exceptional nature of HeT-A and TART is reflected in their Gag proteins, which interact with cellular components in ways different from Gag proteins of other retrotransposons.
Because HeT-A and TART have special roles in the cell, it is not surprising that they are treated differently from other retrotransposons by the cell and, hence, have characteristic localizations. A more unexpected finding of these studies is the element-specific localization seen for the nontelomeric retrotransposons. Two of them, Doc and jockey, share very similar coding sequences, transpose by similar mechanisms, and inhabit much of the same chromosomal regions. Thus, it might be expected that their interactions with the host cell would be very similar, yet the localizations that we see indicate that this is not the case. Their patterns of Gag distribution differ in several ways, suggesting that these proteins undergo multiple interactions as they move to complete the process of transposition. The element-specific localization differences might indicate that Gags of different elements are differentially efficient at negotiating different steps in a shared pathway. Alternatively, they might indicate that different proteins follow slightly different paths.
This study was designed to look specifically for the potential for interactions between Gag and other proteins of the cell in which they transpose. We presume that jockey, Doc, and the two telomeric proteins all transpose in SL2 cells, making those cells appropriate for the study. SL2 cells contain HeT-A, TART, Doc, and jockey elements and have maintained them since the line was established in the early 1970s (13). Maintenance over this time strongly suggests transposition to replenish lost copies. In addition, transcripts of all four elements are detected in the cells used here (K. Traverse, personal communication).
SL2 cells do not appear to contain active I factors. Southern hybridization detected only weak hybridization to I factor probes, presumably because of the defective sequences present in the pericentric regions of all strains (ref. 14; K. Traverse, personal communication). I factors transpose only in the germ-line of females from strains classified as Reactive. Although I factor does not normally inhabit SL2 cells, the cytoplasmic distribution of our GFP-tagged I factor Gag protein is similar to that found by Seleme et al. (6) for a hemagglutinin (HA)-tagged I factor Gag. Their HA-tagged Gag was produced by an I factor undergoing a high rate of autonomous transposition in the germ-line of females of the appropriate genotype. In their study, antibody staining of ovaries showed a very specific pattern of Gag localization. Localization changed as the oocyte developed, but remained cytoplasmic throughout the process of oogenesis. As in all cytological studies, a small amount of nuclear Gag may have escaped detection. Nevertheless, these studies show that, in a situation in which the element is shown to be undergoing a high level of transposition, the bulk of the element's Gag protein remains in the cytoplasm.
Seleme et al. (6) also transiently transfected SL2 cells with a construct bearing the HA-tagged I factor gag ORF expressed from a heat shock promoter. In their experiments, as in ours, the Gag protein remained in the cytoplasm. Thus, the localization in the SL2 cells was cytoplasmic whether the protein was tagged with the HA-epitope or with GFP and whether it was expressed from the hsp70 promoter or the armadillo promoter.
Four of the five Drosophila Gag proteins in this study have both of the motifs that characterize retroviral Gags, zinc knuckles and the major homology region (MHR). All five have CCHC boxes (CX2CX4HX4C), the zinc-binding motif that has been called a zinc knuckle (15) because it forms structures with stubbier projections than those of the C2H2 zinc finger motif (16). Zinc knuckles are very common in retroelement proteins. Unlike the zinc finger motif, they are very rare in cellular proteins (17, 18). Retroviral mutants in Gag zinc knuckles show that these regions have roles in infection as well as their better-known roles in virus assembly (19–21).
The MHR, a 20-aa region found N-terminal of the zinc knuckle region, is the only region with significant homology among different genera of retroviruses. Studies of mutations in the sequence implicate it in many activities in viral assembly as well as infection of new hosts (22–25). This motif also has been found in the Saccharomyces cerevisiae retrotransposon, Ty3. Mutation analysis of the Ty3 MHR also shows that this region is involved in several steps of the transposition process (26). HeT-A, TART, jockey, and Doc also have MHR-like motifs. I factor, which belongs to a distantly related clade (8), does not have this motif.
Both of the common retroelement motifs are found in those parts of the Gag proteins in which there is significant similarity between the different proteins in this study, suggesting that these domains have functions shared by these elements and do not contribute directly to element-specific localization.
The HeT-A and TART Localizations Are Cell Cycle-Related.
Because HeT-A and TART transpose only to chromosome ends, we would expect their protein distribution within the nucleus to reflect the distribution of those ends. The localization of the large Het-dots is consistent in number and intranuclear distribution with our expectation for telomeres. The small Het-dots (which we believe are progressing to their final localization) and the TART dots are too numerous to be correlated completely with chromosome ends, if there is any correlation at all. A direct correlation with telomere sequences is not feasible. Our efforts to surmount this difficulty led to the recognition that the HeT-A structures were cell cycle-associated.
Telomeres can be detected by in situ hybridization to mitotic chromosomes from mutant stocks of Drosophila with abnormally long telomeres but they are not detected reliably on mitotic chromosomes from wild-type flies (27). We have been unable to reliably detect HeT-A or TART sequences on telomeres of mitotic chromosomes in SL2 cells. In addition, there is a significant amount of fragments of HeT-A and TART sequence in pericentric heterochromatin and Y chromosomes (28, 29). Although both of these problems can be overcome when mapping sequences on condensed chromosomes, they make objective analyses of the diffuse interphase chromatin impossible.
Because we could not reliably identify telomere sequences in interphase nuclei, we looked to see whether Gag protein was associated with chromosomes in metaphase. To avoid the possibility that drugs might disrupt an association, we searched through preparations of the randomly cycling cells, identifying metaphase cells by an antibody that recognizes the metaphase-specific form of histone H3. Those analyses did not permit us to deduce the interphase localizations of Het-dots. Instead, they have revealed an unexpected cell cycle-related change in the distribution of both the HeT-A and TART proteins. The structures seen in interphase nuclei are broken down and protein spreads throughout the cell, except over the chromosomes. The cytoplasmic clusters of the other Gags are not affected in mitotic cells, indicating that the change of state, like the localizations, differs for different Gags.
Is the Cytoplasm a Line of Defense Against Retrotransposons?
Although we expected that the telomeric transposon Gags would differ from the nontelomeric Gags in their localization, it was surprising to find that the nontelomeric elements show so much cytoplasmic accumulation. These Drosophila retrotransposons have conserved the two motifs, zinc knuckles and the MHR, found in retroviruses and other retroelements. Studies of other elements implicate these motifs in both nucleic acid binding and in protein–protein associations. Biochemical studies of Gag proteins from two non-LTR retrotransposons, Drosophila I Factor and human LINE-1, have shown those proteins to be capable of nucleic acid chaperone activities such as annealing complementary sequences and promoting strand exchange. These activities might be required in the nucleus during target-primed reverse transcription (30, 31). Thus, what we know about Gag proteins suggests that the successful non-LTR retrotransposon may produce a protein that is designed to efficiently escort the RNA through the cytoplasm into the nucleus, locate the transposition target, and then facilitate reverse transcription. The HeT-A and TART proteins show localizations consistent with this suggestion. In contrast, the other proteins show inefficient progress to a nuclear target.
On the other hand, it has long been recognized that the relationship between cells and their transposable elements is an uneasy truce, with both sides engaged in an evolutionary arms race. Impeding the activities of Gag proteins may be one of the techniques that cells use to keep populations of transposable elements under control. The two known exceptions to the uneasy truce are the telomeric transposons, which are part of the cellular machinery; thus, there is no advantage to limiting their nuclear access. This explanation is supported by the efficient nuclear localization seen for HeT-A and TART Gags. In contrast, the three nontelomeric Gag proteins are all subject to some form of harassment in their movement.
The idea that part of the cellular defense against retrotransposons occurs during cytoplasmic transport could explain results of studies of Gags from some other elements. A few retroelement Gags have been seen in the nucleus as expected. Gag protein of the fission yeast LTR retrotransposon Tf1 localizes to the nucleus (32). When human foamy Virus infects a cell, its Gag moves rapidly to the centriole, apparently positioning itself for entry into the nucleus when the membrane breaks down at metaphase (33). In contrast, studies of the LTR retrotransposon Ty1 from yeast (34), LINE-1 from humans (31), and I factor from Drosophila (6) all have detected Gag only in the cytoplasm. Of course, none of these studies can eliminate the possibility that a small amount of the protein is present in the nucleus. Because transposition of these elements is a rare event, it is possible that the actual entry of Gag, and any associated components of the transposition complex, also is a rare event and therefore not detected against the background of cytoplasmic material.
The study of marked I factor Gag (6) is especially informative because the protein is produced by an element transposing at high rates in the SF strain, with an estimated transposition frequency of about 0.5 transposition events per gamete per generation. Nevertheless, Gag protein appears to be entirely cytoplasmic in that strain. This could indicate that the activity of this protein takes place entirely in the cytoplasm. On the other hand, this protein has been shown to have activities that could be required in the nucleus for the target-primed reverse transcription used for transposition (30). The amount of Gag required in any nucleus to produce 0.5 transposition events in that cell may not be an appreciable fraction of the total protein in the cell. The I factor study provides a second piece of suggestive evidence that changes in cytoplasmic localization may correlate with the success of transposition events. Expression of the marked element in RSF females, where it transposes at lower rates than in the SF females, showed that the protein, still cytoplasmic, had a different pattern of localizations during oocyte development than it had in ovaries, where transposition was occurring frequently. It is possible that this different pattern of localization was a reflection of one way in which transposition is inhibited in RSF flies.
In summary, comparison of the intracellular localization of five Drosophila retrotransposon Gag proteins suggests that, although these proteins may share similar roles in the retrotransposon, their interactions with components of the cell may be different and these interactions may be one of the mechanisms by which the cell influences the level of transposition of an element.
Acknowledgments
We thank H. Biessmann, I. Busseau, T. Heidmann, R. Levis, T. Nguyen, and I. Rebay for their generous gifts of clones. Members of the Pardue laboratory, P. G. DeBaryshe, and Ky Lowenhaupt have provided much useful discussion and helpful comments on the manuscript. This work has been supported by Grants GM57006 and GM50315 from the National Institutes of Health.
Abbreviations
- LTR
long terminal repeat
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- MHR
major homology region
References
- 1.Freed E O. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 2.Whittaker G R, Kann M, Helenius A. Annu Rev Cell Dev Biol. 2000;16:627–651. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- 3.Pardue M-L, DeBaryshe P G. In: Mobile DNA II. Craig N, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am. Soc. Microbiol.; 2002. , in press. [Google Scholar]
- 4.Pardue M-L, DeBaryshe P G. Chromosoma. 1999;108:73–82. doi: 10.1007/s004120050354. [DOI] [PubMed] [Google Scholar]
- 5.Moran J V, Holmes S E, Nass T P, DeBerardinis R J, Boeke J D, Kazazian H H., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 6.Seleme M-D C, Busseau I, Malinsky S, Bucheton A, Teninges D. Genetics. 1999;151:761–771. doi: 10.1093/genetics/151.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardue M-L, Danilevskaya O N, Lowenhaupt K, Wong J, Erby K. J Mol Evol. 1996;43:572–583. doi: 10.1007/BF02202105. [DOI] [PubMed] [Google Scholar]
- 8.Malik H S, Burke W D, Eickbush T H. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 9.Xie W, Gai X, Zhu Y, Zappulla D C, Sternglanz R, Voytas D F. Mol Cell Biol. 2001;21:6606–6614. doi: 10.1128/MCB.21.19.6606-6614.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danilevskaya O N, Petrova D A, Pavlova M A, Koga A, Kurenova E V, Hartl D L. Chromosoma. 1992;102:32–40. doi: 10.1007/BF00352288. [DOI] [PubMed] [Google Scholar]
- 11.Biessmann H, Kasravi B, Bui T, Fujiwara G, Champion L E, Mason J M. Chromosoma. 1994;103:90–98. doi: 10.1007/BF00352317. [DOI] [PubMed] [Google Scholar]
- 12.Danilevskaya O N, Tan C, Wong J, Alibhai M, Pardue M-L. Proc Natl Acad Sci USA. 1998;95:3770–3775. doi: 10.1073/pnas.95.7.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider I. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 14.Crozatier M, Vaury C, Busseau I, Pelisson A, Bucheton A. Nucleic Acids Res. 1988;16:9199–9213. doi: 10.1093/nar/16.19.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summers M F, South T I, Kim B, Hare V. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- 16.Evans R M, Hollenberg S M. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 17.Covey S N. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg J M, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 19.Rein A, Henderson L E, Levin J G. Trends Biochem Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 20.Tanchou V, Decimo D, Pechoux C, Lener D, Rogemond V, Berthoux L, Ottmann M, Darlix J L. J Virol. 1998;72:4442–4447. doi: 10.1128/jvi.72.5.4442-4447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorelick R J, Gagliardi T D, Bosche W J, Wiltrout T A, Coren L V, Chabot D J, Lifson J D, Henderson L E, Arthur L O. Virology. 1999;256:92–104. doi: 10.1006/viro.1999.9629. [DOI] [PubMed] [Google Scholar]
- 22.Strambio-de-Castillia C, Hunter E. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cairns T M, Craven R C. J Virol. 2001;75:242–250. doi: 10.1128/JVI.75.1.242-250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 26.Orlinsky K J, Gu J, Hoyt M, Sandmeyer S, Menees T M. J Virol. 1996;70:3440–3448. doi: 10.1128/jvi.70.6.3440-3448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siriaco G M, Cenci G, Haoudi A, Champion E, Zhou C, Gatti M, Mason J M. Genetics. 2001;160:235–245. doi: 10.1093/genetics/160.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traverse K L, Pardue M L. Chromosoma. 1989;97:261–271. doi: 10.1007/BF00371965. [DOI] [PubMed] [Google Scholar]
- 29.Danilevskaya O, Lofsky A, Kurenova E V, Pardue M-L. Genetics. 1993;134:531–543. doi: 10.1093/genetics/134.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dawson A, Hartswood E, Paterson T, Finnegan D J. EMBO J. 1997;16:4448–4455. doi: 10.1093/emboj/16.14.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin S L, Bushman F D. Mol Cell Biol. 2001;21:467–475. doi: 10.1128/MCB.21.2.467-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang V D, Levin H L. Mol Cell Biol. 2000;20:7798–7812. doi: 10.1128/mcb.20.20.7798-7812.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saib A, Puvion-Dutilleul F, Schmid M, Peries J, de The H. J Virol. 1997;71:1155–1161. doi: 10.1128/jvi.71.2.1155-1161.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenna M A, Brachmann C B, Devine S E, Boeke J D. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]