Abstract
Liver regeneration after partial hepatectomy results primarily from the simple division of mature hepatocytes. However, during embryonic and fetal development or in circumstances under which postnatal hepatocytes are injured, organ regeneration is believed to occur from a compartment of epithelial liver stem or progenitor cells with biliary and hepatocytic bipotentiality. The ability to identify, isolate, and transplant epithelial liver stem cells from fetal liver would greatly facilitate the treatment of hepatic diseases currently requiring orthotopic liver transplantation. Here we report the identification and immortalization by retrovirus-mediated transfer of the simian virus 40 large T antigen gene of primate fetal epithelial liver cells with a dual hepatocytic biliary phenotype. These cells grow indefinitely in vitro and express the liver epithelial cell markers cytokeratins 8/18, the hepatocyte-specific markers albumin and α-fetoprotein, and the biliary-specific markers cytokeratins 7 and 19. Bipotentiality of gene expression was confirmed by clonal analysis initiated from single cells. Endogenous telomerase also is expressed constitutively. After orthotopic transplantation via the portal vein, ≈50% of the injected cells integrated into the liver parenchyma of athymic mice without tumorigenicity. Three weeks after transplantation, cells having seeded in the liver parenchyma expressed both albumin and α-fetoprotein but had lost expression of cytokeratin 19. These results provide strong evidence for the existence of a bipotent epithelial liver stem cell in nonhuman primates. This unlimited source of donor cells also should enable the establishment of a model of allogenic liver cell transplantation in a large animal closely related to humans and shed light on important questions related to liver organogenesis and differentiation.
The liver possesses the unique duality of being composed of cells that are quiescent under normal circumstances and being capable of rapid and extensive regeneration after partial resection (1). Although direct regeneration of human and primate liver from mature adult hepatocytes has been recognized, these cells are spontaneously quiescent in vivo and in vitro (2). With optimized culture conditions in the presence of hepatocyte growth factor, primary adult hepatocytes can undergo only 1–2 population doublings before loosing their differentiation status and dying (3, 4). Only populations of so-called small hepatocytes isolated from adult rats are able to undergo multiple divisions for several weeks, although the nature of these cells remained unclear, and they are not capable of indefinite expansion (5, 6).
These limitations have hampered progress in many areas including the cell and molecular biology of hepatocytes in culture, the study of viruses with liver tropism, and potential applications in allogeneic cell transplantation as a bridge therapy for liver failure. Currently, indefinite expansion of adult hepatocytes in culture can be achieved by transfer of an immortalizing oncogene only (7–9). During fetal liver development in rodents, epithelial liver stem cells have the ability to proliferate moderately in vitro and in vivo and to differentiate along the hepatocytic or bile duct epithelial cell lineage (10). The identification of an epithelial liver stem cell capable of unlimited expansion and bilineage (hepatocyte-biliary cells) differentiation is of great interest for cellular and gene therapies, but its isolation has not been achieved yet in humans and other primates (11–13). The aim of this study was to isolate and immortalize nonhuman primate fetal liver (PFL) stem cells by means of a retroviral vector expressing simian virus 40 large T antigen (SV40T) and to evaluate their homing, functionality, and differentiation into the liver parenchyma after intraportal injection in athymic mice. A clonal population of immortalized cells, referred to as immortalized PFL stem (IPFLS) cells, was obtained and expanded extensively. IPFLS cells displayed a hepatocytic biliary phenotype and were able to engraft in the parenchyma of athymic mice. Three weeks after transplantation, cells having seeded in the liver parenchyma expressed both α-fetoprotein (AFP) and albumin (ALB), markers of the hepatocytic lineage, but had lost expression of cytokeratin 19 (CK19), a bile duct marker.
Materials and Methods
PFL Cells.
Liver from a cynomolgus monkey fetus at a gestational age of 89 days was isolated by standard perfusion and 0.1% collagenase A treatment (Roche Molecular Biochemicals). PFL cells were cryopreserved in heat-inactivated Myoclone super plus FCS (Invitrogen) and 10% dimethyl sulfoxide (Sigma; ref. 14). Liver primary cells were thawed and plated in Primaria multiwell (24-well) tissue culture plates (BD) at a density of 3 × 105 cells per well in DMEM/HAM's F12 medium (Eurobio, Paris) containing 15% FCS and 0.1% bovine serum-ALB (Sigma). The medium was replaced 6 h later and daily thereafter by serum-free DMEM/HAM's F12 medium chemically defined with 10−8 M insulin/10−6 M dexamethasone (Merck Sharp & Dohme)/10−8 M 3,3′-triiodo-l-thyronine/0.24% linoleic acid-ALB (2:1)/1 mg/ml apo-transferrin (iron-poor; Sigma)/100 μg/ml vitamin C (Roche).
Retroviral Vector.
The immortalizing bicistronic retroviral SSR#69 vector (15) was derived from LXSN (a gift from D. Miller, Fred Hutchinson Cancer Research Center, Seattle) and comprises the following elements from 5′ to 3′: a long terminal repeat with packaging signal, a hygromycin-resistant/herpes simplex virus thymidine kinase fusion gene, the encephalomyocarditis virus internal ribosome entry site, the U19 mutant of SV40T, from which the intron was deleted to prevent expression of SV40 small T antigen, and another long terminal repeat preceded by its polypurine tract.
Retroviral Transduction.
The SSR#69 ΨCRIP amphotropic cell line was grown in DMEM (Eurobio) supplemented with 10% heat-inactivated newborn calf serum (Invitrogen)/10−6 M dexamethasone/5 mM n-butyric acid sodium salt (Sigma) to increase viral titers as described (16). Viral titers were up to 106 particle-forming units per ml as estimated by viral RNA slot blot analysis using a LacZ virus as a control (data not shown). PFL cells were incubated with 0.5 ml of viral supernatant containing 3 μg/ml polybrene (Sigma) per Primaria multiwell (24-well) tissue-culture plate for 4 h (multiplicity of infection = 10) at days 2 and 3 postplating. Cells then were cultured in chemically defined DMEM/HAM's F12/William's E medium (1:1:2) containing 5% Myoclone Super plus FCS.
Southern Blot Analysis.
Genomic DNAs were digested overnight with SacI (Biolabs, Northbrook, IL), which cuts only once in each long terminal repeat. Digested DNA (20 μg) was separated by electrophoresis on 1% agarose gel, transferred onto a nitrocellulose membrane, and hybridized with a 32P-labeled SV40T probe.
Limited Cell Dilution for Clonal Analysis.
IPFLS cells were trypsinized and seeded in several dishes at a density of 200,000 cells per 10-mm dish. Cells were grown for 1 week with medium changes every other day, and ≈100 clones were subsequently analyzed by double immunofluorescence staining for both ALB and CK19.
Immunohistochemistry and Immunocytochemistry.
IPFLS cells first were treated with PBS/4% formaldehyde followed by PBS/0.1% Triton for 5 min before incubating them with either a mouse monoclonal IgG2a anti-SV40T antibody (Pab 101, 1/1,000) or a goat polyclonal anti-p53 antibody (N-19, 1/200; Santa Cruz Biotechnology). Liver biopsies from fetal monkeys and nude mice were cryopreserved in isopentane immersed in liquid nitrogen and stored at −80°C. Cryostat tissue sections of 7 μm in thickness and cultured PFL and IPFLS cells were fixed in methanol/acetone (1:1) and then pretreated with PBS containing 1% gelatin. Fetal monkey tissues and cells were incubated with mouse monoclonal IgG anti-CK7 (LP1K), anti-CK8/18 (LE61; a gift from B. Lane, Dundee University, Dundee, Scotland), or anti-CK19 (1/100; Dako) antibodies for 1 h at room temperature. To reveal fetal liver markers, fetal monkey or transplanted athymic mice tissues and cells were incubated with mouse monoclonal IgG anti-ALB (CL2513A, 1/50; Cedarlane Laboratories), anti-AFP (C3, 1/50; Santa Cruz Biotechnology), or anti-CK19 (1/100; Dako) antibodies for 1 h at room temperature. These mouse antibodies recognize the corresponding simian ALB, AFP, or CK19 but not the mouse proteins. Bound goat antibodies were detected with FITC-labeled rabbit polyclonal anti-goat IgG antibodies (1/200; Dako). Bound murine antibodies were detected with secondary polyclonal goat anti-mouse IgG antibodies labeled with horseradish peroxidase (1/50), Cy3 (1/400; Amersham Pharmacia), or FITC (1/200; Santa Cruz Biotechnology). For double labeling of ALB and CK19, IPFLS cells were incubated for 1 h with mouse monoclonal anti-human CK19 (Dako) followed by Cy3-conjugated anti-mouse IgG (Amersham Pharmacia) and FITC-conjugated monoclonal anti-human ALB antibodies. Counterstaining of nuclei was performed with either hematoxylin for peroxidase staining or 4′,6-diamidino-2-phenylindole (Vector Laboratories) for fluorescence staining.
Telomerase Repeat Amplification Protocol Assay.
Telomerase activity was assayed on lysates of IPFLS cells and fetal liver cells cultured for 1, 2, and 3 days by using the TRAPeze PCR kit (Oncor). PCR products were analyzed by the ELISA method (data not shown) and migration on agarose gel electrophoresis.
Tumorigenicity Assay.
Tens of millions of cells were injected s.c. into the flanks of 4–5-week-old male nude mice. Animals were examined weekly for the presence of tumors during a 3-month period.
IPFLS Cell Transplantation.
Eight male nude mice underwent 50% hepatectomy 40 h before cell infusion. IPFLS cells (1,000,000) were injected via the portal vein into the liver of six hepatectomized athymic mice. PBS (1×) was injected in the portal vein of two animals as a control. Liver biopsies were performed 7 and 21 days posttransplantation. Eight male alymphoid RAG2−/−/γc−/− mice underwent the same protocol and were killed 50 days after transplantation to evaluate the tumorigenic potential of IPFLS cells.
Quantification of the Degree of Cell Chimerism.
We infused 106 hepatocytes via the portal vein, which represents ≈1% of the mouse hepatocytic mass, because it has been estimated that 1 g of liver contains ≈108 hepatocytes. ALB-immunostained cells were counted in all transplanted mouse livers from a total of 150 microscopic fields. Assuming that 450 hepatocytes can be visualized per microscopic field under the same conditions (7-μm liver section; magnification, ×200), the degree of cell chimerism with donor cells was calculated as follows: observed percentage of cell chimerism = [number of transplanted cells counted/150)/450] * 100.
Results
Immunohistochemistery of Nonhuman PFL.
We examined frozen sections of cynomolgus monkey fetal liver (2/3 gestation) by immunohistochemistry. We analyzed well established lineage-specific markers: ALB and AFP as hepatocyte-specific markers, CK7 and CK19 as biliary-specific markers, and CK8/18 as a general liver epithelial cell marker.
The PFL was composed of epithelial cells interspersed with islets of nonstaining hematopoietic cells (Fig. 1A, arrows). Epithelial cells with a phenotype of hepatocyte (ALB+, AFP+, CK8/18+) were dispersed throughout the parenchyma (Fig. 1 B–D). Epithelial cells with the biliary phenotype (CK7+, CK19+) formed large biliary ducts (Fig. 1 E and F) and also appeared to be localized at the margin of the primitive portal tracts (Fig. 1 G and H). Because epithelial liver stem cells are expected to represent a rare subset of cells within the liver parenchyma, they could not be distinguished unambiguously on frozen sections from these two more predominant cell types.
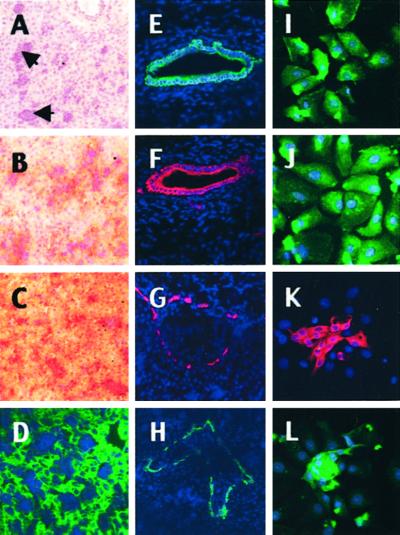
Figure 1.
Immunohistochemistry of cynomolgus monkey fetal liver for lineage-specific markers. (A–H) Liver sections (2/3 gestation). (A) Hematoxylin staining: most of the parenchyma is occupied by epithelial cells interspersed with islets of hematopoietic cells (arrows). (B–H) Immunohistochemistry with monoclonal antibodies that recognize specific simian markers as described in Materials and Methods. Two populations of epithelial cells coexist. The first type has the phenotype of hepatocytes coexpressing ALB (B, peroxidase staining and nuclear counterstaining with hematoxylin), AFP (C), and CK8/18 (D, FITC labeling and nuclear counterstaining with 4′,6-diamidino-2-phenylindole). The other type has a phenotype of biliary cells and/or progenitor cells that coexpress CK7 (E and G, Cy3 labeling and nuclear counterstaining with 4′,6-diamidino-2-phenylindole) and CK19 (F and H) in large biliary ducts and at the margin of the primitive portal tracts. Magnification, ×100. (I–L) Immunocytochemistry of freshly dissociated PFL cells (2/3 gestation). After cell dissociation, most cultured liver cells exhibit a phenotype of hepatocytes expressing ALB (I), AFP (J); clusters of proliferating cells exhibit a phenotype of biliary cells and/or progenitors cells expressing CK7 (K) and CK19 (L). Control cells and tissues did not stain specifically (data not shown). Magnification, ×400.
Cell Composition of Dissociated PFL.
For further analysis, the entire liver of an 89-day-old monkey fetus was perfused through the umbilical vein by using the “two-step” collagenase method. After cell dissociation, the yield of viable PFL cells was 1.35 × 108. PFL cells were cryopreserved as described (14). After thawing, cell recovery was estimated to be 75%, and cells exhibited a morphology similar to that of freshly isolated PFL cells (data not shown). Thawed, cryopreserved, dissociated PFL cells were cultured for 3 days and subsequently analyzed by immunocytochemistry with the same panel of antibodies, revealing the following staining pattern. The most abundant subset of cells expressed markers of fetal hepatocytes (ALB+, AFP+; Fig. 1 I and J), whereas the other expressed markers present on either liver biliary cells or liver stem cells (CK7+, CK19+; Fig. 1 K and L). Here again, the strong predominance of fetal hepatocytes prevented the identification of the rare epithelial liver stem cells by immunohistochemistry. We also observed that at the end of the 3-day culture period, the subset of cells that expressed biliary-specific markers formed tightly organized clusters of ≈16 cells, in contrast to the fetal hepatocytes. This finding suggested that these cells underwent an average of three cell divisions during the 3-day period of in vitro culture instead of a single cell division for fetal hepatocytes.
Immortalization of Fetal Primate Liver Stem Cells.
We defined conditions under which 90% of thawed fetal liver cells were transduced by using a retrovirus encoding the Escherichia coli LacZ gene (data not shown). Then we made use of a Moloney murine leukemia virus vector expressing the immortalizing SV40T, referred to as SSR#69 (ref. 11; Fig. 2A) as follows: 1.8 × 107 PFL cells were transduced twice 48 and 65 h postplating by exposure to amphotropic retroviral supernatants. Nontransduced PFL cells died within 2 weeks, whereas transduced clones arose within 4 weeks. After 6 months, 3 of the 144 emerging clones were still proliferating, and after 12 months, 2 of these clones underwent growth arrest and died (Fig. 2B). The surviving clone, referred to as IPFLS cells, was composed of cells with a cuboidal shape and mononucleated morphology (Fig. 2C), which are characteristics of primary fetal liver cells (Fig. 2D). IPFLS cells grew as a continuous cell line in vitro and were amplified and cryopreserved efficiently.
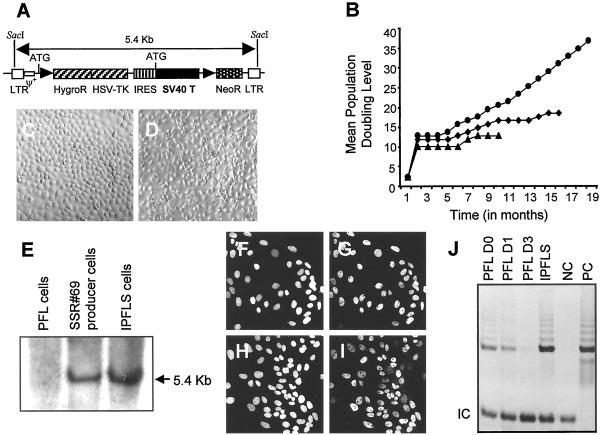
Figure 2.
Immortalization of the PFL stem cell. (A) Structure of the immortalizing retroviral vector. LTR, long terminal repeat; HygroR, hygromycin resistance; HSV-TK, herpes simplex virus/thymidine kinase; IRES, internal ribosome entry site.(B) Growth curve of the three surviving transduced clones: PFL6 (▴), PFL1 (♦), and IPFLS (●). (C and D) Morphology of IPFLS and PFL cells, respectively. (E) Southern blot analyses for provirus integration in PFL, SSR#69 producer, and IPFLS cells. The nuclear expression of SV40T antigen (G) and the protein p53 (I) in IPFLS cells colocalized with nuclear counterstaining with 4′,6-diamidino-2-phenylindole (F and H). (J) Telomerase activity was detected in PFL cells 1 day after thawing but became undetectable after 3 days of culture; in contrast, telomerase activity was constitutively expressed in IPFLS cells. PC and NC, positive and negative controls, respectively.
Characteristics of IPFLS Cells.
We analyzed the growth kinetics of IPFLS cells by measuring the mean population-doubling levels. To date, cells have been passaged for 30 months, which corresponds to 75 mean population-doubling levels with an average of one population doubling every 6 days. Southern blot analysis of IPFLS cells demonstrated stable proviral integration (Fig. 2E). To determine whether the SV40T gene was translated effectively into protein, we assessed its expression in IPFLS cells by immunocytochemistry. All cells expressed SV40T in their nuclei (Fig. 2 F and G). In addition, nuclear p53, which is undetectable in normal cells, also was present (Fig. 2 H and I). SV40T is known to bind to p53, thereby preventing the p53-dependent cell cycle arrest occurring during senescence (17). We also assessed telomerase activity in IPFLS cells by using the telomere repeat amplification protocol and compared it to that found in PFL cells (18–20). As expected for fetal tissues, telomerase activity was detectable in cryopreserved PFL cells. It decreased after 24 h of culture and became undetectable 72 h after plating. By contrast, IPFLS cells displayed a high level of telomerase activity (Fig. 2J).
To determine the phenotypic identity of IPFLS cells, we analyzed the expression of lineage-specific markers by immunocytochemistry. All IPFLS cells expressed hepatocytic (ALB+, AFP+, CK8/18+) and biliary (CK7+, CK19+; Fig. 3 A–E) cell markers. We also performed clonogenic assays in which a single cell was expanded to reach a cluster of proliferating cells to analyze the expression of both hepatocytic and biliary markers within the same cells. All cells within these clusters expressed both ALB and CK19 without apparent heterogeneity or variegation (Fig. 3 G and H), thereby demonstrating their bipotency.
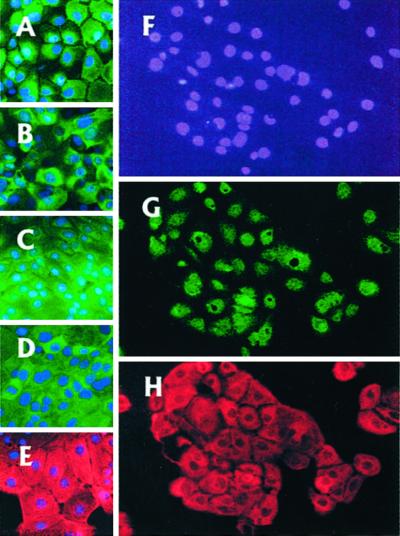
Figure 3.
Characterization of the PFL stem cell. IPFLS express ALB, AFP, CK8/18, CK7, and CK19 (A–E, respectively). (F–H) Bipotent nature of IPFLS cells. After 7 days of culture, each single IPFLS cell proliferated to reach a cluster that express both ALB (G) and CK19 (H). Magnification, ×400.
IPFLS Injection into Athymic Mice.
We first investigated the tumorigenic potential of IPFLS cells in athymic nude mice (21). When nude mice were injected s.c. with 107 IPFLS cells, they did not develop any tumor for as long as 16 weeks after infusion, whereas mice injected with the same amount of HuH7 human hepatoma cells developed tumors 3 weeks after cell transplantation (data not shown). Next, we investigated whether IPFLS cells have the ability to integrate and function within the adult liver parenchyma. We infused 1 × 106 IPFLS cells (1% of the murine hepatocytic mass) via the portal vein into the liver of athymic nude mice. We assessed the degree of hepatic engraftment achieved 7 days after transplantation by immunohistostaining of liver biopsies with a monoclonal antibody that recognizes simian but not murine ALB. After injection of ≈1% of the total number of hepatocytes present in a normal mouse liver, 28.6–86.9% of injected simian cells, depending on the animal, were observed in the liver parenchyma 7 days after transplantation (Table 1). Transplanted cells were scattered throughout periportal areas and around the portal vein (Fig. 4A), resulting in a mean chimerism of 0.47% of the total number of hepatocytes present in a normal mouse liver. The fate of the injected cells was determined also by a second biopsy 21 days after transplantation. The transplanted cells were found to be integrated into the liver parenchyma close to host hepatocytes (Fig. 4B). Some of them exhibited two nuclei, a feature characteristic of adult hepatocytes (Fig. 4C). We also analyzed the coexpression of fetal hepatic markers on serial liver sections with specific antibodies. The same IPFLS cells coexpressed both ALB (Fig. 4 D and E) and AFP (Fig. 4 F and G) but not CK19 (Fig. 4 H and I). Livers of control mice did not stain with the simian-specific ALB, AFP, and CK19 antibodies, whereas control simian fetal livers did (data not shown). Finally, potential tumorigenicity was evaluated further in alymphoid RAG2−/−/γc−/− mice after intraportal injection of 106 IPFLS cells (22). No tumors in the liver or other sites were detected after 7 weeks (data not shown).
Table 1.
Degree of cell chimerism in mice transplanted with IPFLS cells
| Mouse | Theorical cell chimerism, % | Number of engrafted cells (150 fields) | Observed cell chimerism, % | Transplanted cells engrafted, % | Transplanted cells lost, % |
|---|---|---|---|---|---|
| I (Control) | 0 | ||||
| II | 1 | 261 | 0.386 | 38.6 | 61.4 |
| III | 1 | 0 | 0.000 | 0.0 | 100.0 |
| IV | 1 | 587 | 0.869 | 86.9 | 13.1 |
| V (Control) | 0 | ||||
| VI | 1 | 391 | 0.579 | 57.9 | 42.1 |
| VII | 1 | 496 | 0.727 | 72.7 | 27.3 |
| VIII | 1 | 193 | 0.286 | 28.6 | 71.4 |
| Mean | 321 | 0.475 | 47.5 | 52.6 |
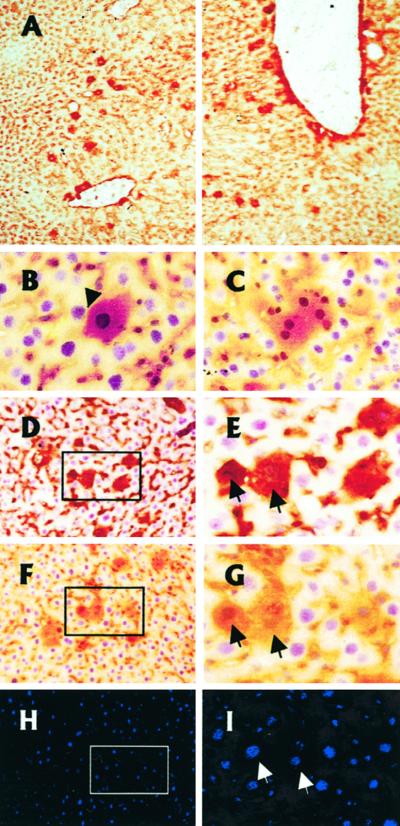
Figure 4.
Functionality of IPFLS cells after xenotransplantation in adult athymic mouse liver. IPFLS cells (1 × 106) were infused via the portal vein. We visualized hepatic engraftment and differentiation by immunohistochemistry of serial frozen liver sections with monoclonal mouse antibodies that recognize simian but not mouse ALB, AFP, or CK19. (A) Liver sections were performed 8 days after transplantation and stained for simian ALB. Transplanted IPFLS cells were scattered throughout periportal areas and in the vicinity of the portal tracts. Magnification, ×100. (B–I) Liver sections were performed 21 days after transplantation. (B–G) Peroxidase staining for ALB and nuclear counterstaining with hematoxylin. (B and C) Transplanted cells have entered the hepatic parenchyma (arrow), and some of them exhibit two nuclei, a feature characteristic of adult hepatocytes. Magnification, ×400. (D–I) Immunostaining performed on serial liver section shows the differentiation pattern of the same transplanted cells (arrows). (D and E) Section immunostained for simian AFP. (F–I) Section immunostained for simian ALB (F and G, peroxidase staining) and for simian CK19 (H and I, Cy3 labeling). Magnification, ×200 and ×400.
Discussion
Based on data reported previously in rodents, the putative primate epithelial liver stem cells should share markers of both hepatocytic and biliary lineages and be localized within primitive ductal structures (23–27). To document the existence of such cells as well as the cell-type composition and organization of nonhuman PFL, we examined frozen sections of monkey fetal liver (2/3 gestation) by using lineage-specific markers. The PFL was composed of epithelial cells interspersed with islets of nonstaining hematopoietic cells. Epithelial cells with a phenotype of hepatocyte were dispersed throughout the parenchyma, whereas epithelial cells with the biliary phenotype formed large biliary ducts and also appeared to be localized at the margin of the primitive portal tracts. Immunocytochemistry analysis of PFL cells from an entire liver of a monkey fetus (89-day-old) revealed that the most abundant subset of cells expressed markers of fetal hepatocytes, whereas the other expressed markers present on either liver biliary cells or liver stem cells. Furthermore, we found that a subset of cells expressing biliary-specific markers underwent a greater number of cell divisions in vitro, forming tightly organized clusters. We hypothesized that this minor subset of cells may contain the putative epithelial liver stem cells and be transduced preferentially by a Moloney murine leukemia virus-based retroviral vector, the infectivity of which is restricted to cells in active division.
By means of a Moloney murine leukemia virus retroviral vector expressing SV40T, we isolated a transduced clone, referred to as IPFLS, that grows as a continuous cell line in vitro and is amplified and cryopreserved efficiently. Southern blot analysis of transduced IPFLS cells demonstrated stable proviral integration. Immunocytochemistry analysis for SV40T and p53 showed concordant expression in the nucleus as expected from SV40T-mediated immortalization of mammalian cells. IPFLS cells showed a bipotent hepatocytic and biliary phenotype without apparent heterogeneity or variegation detected by immunocytochemistry. Hence, IPFLS cells harbor a phenotype expected from the primate epithelial liver stem cell.
Because constitutive expression of telomerase has been linked to the immortalization process, we assessed telomerase activity in IPFLS and PFL cells (18–20). IPFLS cells displayed a high level of telomerase activity in contrast to proliferating PFL cells 3 days after plating. Therefore, telomerase activity was not associated with in vitro proliferation, in contrast to what was suggested for other cell types and species (28), but with the immortalization process of the primate epithelial liver stem cell. Although the 16-cell cluster formation of cells that expressed biliary-specific markers indicated that many of these cells initially were dividing in vitro and therefore were transducible by the high-titer Moloney murine leukemia virus-based retroviral vector, the frequency of PFL cell immortalization was only 10−7. The fact that the truly immortal clone that emerged, IPFLS, constitutively expresses telomerase and has a phenotype of stem cell is consistent with the hypothesis that two conditions may have been required for successful immortalization and that only the liver stem cell would have met both criteria: initial division in culture and spontaneous constitutive expression of telomerase.
In vitro long-term cultures of IPFLS cells still respond to contact inhibition at confluence and do not exhibit morphological criteria of transformation. In vivo, when nude mice were injected s.c. with IPFLS cells or in the portal vein of alymphoid RAG2−/−/γc−/− mice, they did not develop any tumors. We then assessed the degree of hepatic engraftment achieved 8 days after intraportal injection of IPFLS cells in athymic mice. We observed that injected IPFLS cells were scattered throughout periportal areas and around the portal vein. We calculated that ≈50% of the transplanted cells had engrafted, which is consistent with reports of syngeneic transplantation of rat hepatocytes (29). The expression of biliary and hepatic markers was subsequently analyzed in vivo. After 21 days, IPFLS cells that seeded into the liver parenchyma coexpressed both simian ALB and AFP but not CK19, thereby demonstrating that in our xenotransplantation mouse model, injected cells remained functional in vivo and differentiated into the hepatocyte lineage. In addition we observed several engrafted cells with two nuclei, a feature characteristic of adult hepatocytes. Conversely, it remains to be determined whether IPFLS cells seeding in the vicinity of biliary areas would differentiate into the biliary lineage and lose expression of hepatocytic markers. The rarity of this event precluded us to draw a conclusion at this point.
It has been reported previously that in rodents the liver can be repopulated by genetically engineered hepatocytes harboring a selective advantage over resident hepatocytes (30–32). After transplantation into syngeneic adult rat liver, primary rat fetal liver progenitor cells also were able to engraft and proliferate (33–34). More recently, in a mouse model of hereditary tyrosinemia type 1, the FAH−/− mouse liver was repopulated with purified hematopoietic stem cells indicating that in this model hematopoietic stem cells can differentiate into hepatocytes (35).
Our data suggest that we now have immortalized a subset of liver cells successfully with a phenotype expected from the putative primate epithelial liver stem cell. Unlike primary hepatocytes, these cells grow indefinitely in vitro, can be cryopreserved, and remain phenotypically stable. These cells can engraft and remain functional in athymic mouse liver without tumorigenicity, thus rendering the reversible immortalization method we reported previously more dispensable (36). Although the differentiation pattern and the fate of these cells need to be explored further in mouse models of metabolic disease, our results suggest that the immortalized IPFLS primate epithelial liver stem cells would permit various basic studies of this rare cell type and allow the development of preclinical protocols of allotransplantation in a primate model closely related to humans.
Acknowledgments
We thank F. Capron for helpful advice, Jacqueline Jouanneau for assistance with the athymic mice, James Di Santo for the RAG2−/−/γc−/− mice, François Boussin and Laurent Gauthier for the telomerase repeat amplification protocol assay, and Myriam Bennoun and Isabelle Bouchaert for technical support. We acknowledge the Department of Primate Research at the Institut National pour la Recherche Agronomique. This research was supported in part by grants from the Association Française contre les Myopathies, the Association pour la Recherche contre le Cancer, and the Etablissement Français des Greffes (to A.W.) and National Institutes of Health Grants HL55435 and DK56174 (to P.L.). J.-E.A. was supported by the Ministère de la Recherche and the Fondation pour la Recherche Médicale. D.M.-C. and N.L. were supported by the Institut National de la Santé Et de la Recherche Médicale and the Association Française contre les Myopathies, respectively.
Abbreviations
- SV40T
simian virus 40 large T antigen
- PFL
primate fetal liver
- IPFLS
immortalized PFL stem
- AFP
α-fetoprotein
- ALB
albumin
- CK
cytokeratin
References
- 1.Michalopoulos G K, DeFrances M C. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 2.Fausto N. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 3.Matsumoto K, Nakamura T. In: Hepatocyte Growth Factor (HGF-SF) and the c-met Receptor. Goldberg I D, Rosen E M, editors. Basel: Birkhauser; 1993. pp. 225–249. [Google Scholar]
- 4.Runge D, Runge D M, Jager D, Lubecki K A, Beer Stolz D, Karathanasis S, Kietzmann T, Strom S C, Jungermann K, Fleig W E, Michalopoulos G K. Biochem Biophys Res Commun. 2000;269:46–53. doi: 10.1006/bbrc.2000.2215. [DOI] [PubMed] [Google Scholar]
- 5.Tateno C, Takai-Kajihara K, Yamasaki C, Sato H, Yoshizato K. Hepatology. 2000;31:65–74. doi: 10.1002/hep.510310113. [DOI] [PubMed] [Google Scholar]
- 6.Katayama S, Tateno C, Asahara T, Yoshizato K. Am J Pathol. 2001;158:97–105. doi: 10.1016/S0002-9440(10)63948-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodworth C D, Kreider J W, Mengel L, Miller T, Meng Y L, Isom H C. Mol Cell Biol. 1988;10:4492–4501. doi: 10.1128/mcb.8.10.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeifer A M A, Cole K E, Smoot D T, Weston A, Groopman J D, Shields P G, Vignaud J M, Juillerat M, Lipsky M M, Trump B F, Lechner J F, Harris C C. Proc Natl Acad Sci USA. 1993;90:5123–5127. doi: 10.1073/pnas.90.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amicone L, Spagnoli F M, Spath G, Giordano S, Tommasini C, Bernardini S, De Luca V, Della Rocca C, Weiss M C, Comoglio P M, Tripodi M. EMBO J. 1997;16:495–503. doi: 10.1093/emboj/16.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germain L, Blouin M J, Marceau N. Cancer Res. 1988;48:4909–4918. [PubMed] [Google Scholar]
- 11.Kubota H, Reid L M. Proc Natl Acad Sci USA. 2000;97:12132–12137. doi: 10.1073/pnas.97.22.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann U, Crosby H A, Ramani P, Kelly D A, Strain A J. Hepatology. 1999;30:112–117. doi: 10.1002/hep.510300140. [DOI] [PubMed] [Google Scholar]
- 13.Andreoletti M, Pages J C, Mahieu D, Loux N, Farge D, Sacquin P, Simon L, Hamza J, Bargy F, Briand P, Leperq J, Weber A. Hum Gene Ther. 1997;8:267–274. doi: 10.1089/hum.1997.8.3-267. [DOI] [PubMed] [Google Scholar]
- 14.Haruna Y, Saito K, Spaulding S, Nalesnik M A, Gerber M A. Hepatology. 1996;23:476–481. doi: 10.1002/hep.510230312. [DOI] [PubMed] [Google Scholar]
- 15.Westerman K, LeBoulch P. Proc Natl Acad Sci USA. 1996;93:8971–8976. doi: 10.1073/pnas.93.17.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pages J C, Loux N, Farge D, Briand P, Weber A. Gene Ther. 1995;2:547–551. [PubMed] [Google Scholar]
- 17.Lane D P, Crawford L V. Nature (London) 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 18.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 19.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, Wang H, Bishop J M, Blackburn E H. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 22.Cooper R N, Irintchev A, Di Santo J P, Zweyer M, Morgan J E, Partridge T A, Butler-Browne G S, Mouly V, Wernig A. Hum Gene Ther. 2001;12:823–831. doi: 10.1089/104303401750148784. [DOI] [PubMed] [Google Scholar]
- 23.Shiojiri N, Lemire J M, Fausto N. Cancer Res. 1991;51:2611–2620. [PubMed] [Google Scholar]
- 24.Grisham J W, Thorgeirsson S S. In: Stem Cells. Potten C S, editor. Orlando, FL: Academic; 1997. pp. 233–282. [Google Scholar]
- 25.Alison M. Curr Opin Cell Biol. 1998;10:710–715. doi: 10.1016/s0955-0674(98)80111-7. [DOI] [PubMed] [Google Scholar]
- 26.Strain A J, Crosby H A. Gut. 2000;46:743–745. doi: 10.1136/gut.46.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schafritz D A. Hepatology. 2000;32:1399–1400. doi: 10.1053/jhep.2000.20818. [DOI] [PubMed] [Google Scholar]
- 28.Greider C W. Proc Natl Acad Sci USA. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta S, Rajvanshi P, Sokhi R P, Vaidya S, Irani A N, Gorla G R. Hepatology. 1999;29:509–519. doi: 10.1002/hep.510290213. [DOI] [PubMed] [Google Scholar]
- 30.Rhim J A, Sandgren E P, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1995;92:4942–4946. doi: 10.1073/pnas.92.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Overturf K, Al-Dhalimy M, Tanguay R, Brantly M, Ou C N, Finegold M, Grompe M. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 32.Mignon A, Guidotti J E, Mitchell C, Fabre M, Wernet A, De La Coste A, Soubrane O, Gilgenkrantz H, Kahn A. Nat Med. 1998;4:1185–1188. doi: 10.1038/2681. [DOI] [PubMed] [Google Scholar]
- 33.Dabeva M D, Petkov P M, Sandhu J, Oren R, Laconi E, Hurston E, Shafritz D A. Am J Pathol. 2000;156:2017–2031. doi: 10.1016/S0002-9440(10)65074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandhu J S, Petkov P M, Dabeva M D, Shafritz D A. Am J Pathol. 2001;159:1323–1334. doi: 10.1016/S0002-9440(10)62519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagasse E, Connors H, Al-Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman I L, Grompe M. Nat Med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi N, Fujiwara T, Westerman K A, Inoue Y, Sakaguchi M, Noguchi H, Miyazaki M, Cai J, Tanaka N, Fox I J, Leboulch P. Science. 2000;287:1258–1262. doi: 10.1126/science.287.5456.1258. [DOI] [PubMed] [Google Scholar]