Abstract
Expression of tobacco mosaic virus (TMV) coat protein (CP) in plants confers resistance to infection by TMV and related tobamoviruses. Certain mutants of the CP (CPT42W) provide much greater levels of resistance than wild-type (wt) CP. In the present work, infection induced by RNA transcripts of TMV clones that contain wt CP or mutant CPT42W fused to the green fluorescent protein (GFP) (TMV-CP:GFP, TMV-CPT42W:GFP) and clones harboring TMV movement protein (MP):GFP were followed in nontransgenic and transgenic tobacco BY-2 protoplasts and Nicotiana tabaccum Xanthi-nn plants that express wt CP or CPT42W. On nontransgenic and wt CP transgenic plants, TMV-CP:GFP produced expanding, highly fluorescent disk-shaped areas. On plants expressing CPT42W, infection by TMV-CP:GFP or TMV-MP:GFP-CP produced infection sites of smaller size that were characterized by low fluorescence, reflecting reduced levels of virus spread and reduced accumulation of both CP:GFP and MP:GFP. TMV-CPT42W:GFP failed to produce visible infection sites on nontransgenic plants, yet produced normal infection sites on MP-transgenic plants that produce MP. TMV infection of transgenic BY-CPT42W protoplasts resulted in very low levels of MP accumulation, whereas on BY-CP protoplasts (containing wt CP), infection produced higher levels of MP than in nontransgenic BY-2 cells. The results suggest that wt CP has a positive effect on the production of MP, whereas the CPT42W has a negative effect on MP accumulation and/or function. This effect results in very high levels of resistance to TMV infection in plants containing CPT42W. This report shows that the CP of a plant virus regulates production of the MP, and that a mutant CP interferes with MP accumulation and cell-to-cell movement of infection.
Coat protein (CP)-mediated resistance (CP-MR) refers to resistance of transgenic plants that produce virus CP to infections against the virus from which the CP is taken (1). Studies of CP-MR against tobacco mosaic virus (TMV) have increased the understanding of the mechanisms that govern CP-MR (for review, see refs. 2 and 3). As a result of such studies, it is known that transgenic CP interferes with virus disassembly early after virus entry (3–5). Furthermore, mutant CPs that lack the ability to self-aggregate failed to protect against TMV infection, whereas mutants with increased CP–CP interactions provided higher levels of protection than did wild-type (wt) CP (6). These studies established a direct correlation between the degree of CP aggregation and the levels of protection against TMV infection and shed light on its role in blocking virus disassembly (6). They also suggested that certain mutants of CP affected either virus replication and/or cell-cell spread of infection.
TMV RNA encodes an RNA-dependent RNA polymerase (7) and the 30-kDa movement protein (MP). MP is dispensable for virus replication but essential for cell-to-cell spread (8, 9). The CP is dispensable for virus replication and cell-to-cell movement (10–13) and is essential for rapid long-distance movement of TMV in plants (12, 14). The goal of the present study was to elucidate the role of a specific CP mutant in which amino acid 42 was mutated from threonine to tryptophan (CPT42W) and restricted TMV infection (6).
In recent years, the green fluorescent protein (GFP) from the jellyfish Aequorea victoria (15) has been widely used as a marker to study the cellular and molecular aspects of virus replication, cell-to-cell spread of infection, and the interactions between viral proteins and host components. Fusion of GFP to viral proteins has led to new insights into the temporal and spatial distribution of gene products, their function(s), and the mechanisms by which they act, especially in elucidating the mechanisms of virus movement in infected plants (for review, see ref. 16).
To understand the cellular mechanisms of CP-MR in plants expressing wt CP and in highly resistant plants expressing the CPT42W mutant, we followed TMV infection through fluorescence and confocal microscopy of GFP-fusion proteins. The spatial and temporal subcellular distribution and levels of accumulation of CP and MP were followed in infected nontransgenic and transgenic plants and BY-2 cells expressing wt CP or CPT42W. The results show that CPT42W, but not wt CP, modulates virus movement by reducing the accumulation and/or function of MP. The implication of the results for virus movement, replication, and the mechanisms of CP-MR are discussed.
Material and Methods
Plant Lines and BY-2 Transgenic Line.
Transgenic lines of Nicotiana tabaccum Xanthi-nn that produce wt TMV CP (line 3646; ref. 1), CPT42W (line nn T42W; ref. 6), or MP (line 277; ref. 8) were homozygous for the transgene. Nontransgenic N. tabaccum and transgenic plants were grown under standard greenhouse conditions or in growth chambers maintained at 24° to 26°C, 14 h light/10 h dark cycle.
Transgenic BY-2 cell lines that produce CPT42W or wt CP were generated via Agrobacterium-mediated transformation by using the clones pMONCP-T42W (6) and pCGN-CPU1 (see below).
Plasmid Constructs.
The clones that encode TMV-MP:GFP, which produces MP:GFP fusion protein, and TMV-ΔC-GFP, which produces the GFP as free protein from the CP subgenomic promoter, are described in Heinlein et al. (17). The clone that encodes TMV-MP:GFP-CP contains the MP fused to the GFP plus the CP, as described in Oparka et al. (18). TMV-T42W containing the Thr-42 of the CP mutated to Trp is described in Bendahmane et al. (6).
Construction of TMV-CP:GFP.
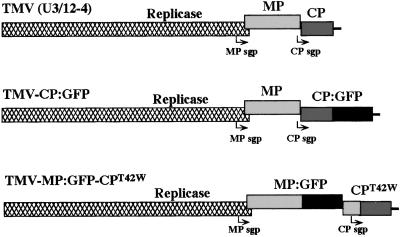
The coding sequence of GFP was fused in-frame to the sequence encoding TMV CP following amino acid Ser-154 by using the vector pCPs/p (19) as follows. The DNA fragment NcoI–BamHI (filled in by Klenow polymerase) containing the full-length GFP ORF was recovered from the plasmid pCKGFPS65T (20, 21) and ligated with the vector pCPs/p previously linearized with SpeI (filled in with Klenow) and NsiI (treated with the T4 DNA polymerase to remove the 3′ overhang). The resulting construct, referred to as pKNcp:gfp, encodes the CP fused in-frame with GFP. The fragment NcoI–KpnI, containing the CP:GFP gene fusion then was excised from the plasmid and inserted into the full-length TMV cDNA U3/12–4 (22) previously linearized with NcoI plus KpnI. The resulting construct is a full-length cDNA clone referred to as pTMV-CP:GFP (Fig. 1). Infection by RNA transcribed from this clone and other CP:GFP fusion proteins (below) did not result in accumulation of virus particles.
Figure 1.
Schematic representation of TMV clones used in this study. MP, movement protein; CP, coat protein; MP sgp, MP subgenomic promoter; CP sgp, coat protein subgenomic promoter; GFP, green fluorescent protein.
Construction of TMV-CPT42W:GFP.
The DNA fragment AccI–KpnI contains the C-terminal part of the CP:GFP gene fusion and the DNA fragment corresponding to the noncoding 3′ end of TMV RNA. This fragment was recovered from the clone pKNcp:gfp after digestion with KpnI and partial digestion with AccI (there are two AccI restriction sites in the GFP sequence). The appropriate AccI–KpnI fragment was inserted in the cDNA clones pTMV-T42W (6) to yield the construct pTMV-CPT42W:GFP (Fig. 1).
Construction of TMV-MP:GFP-CPT42W.
The Thr residue at amino acid 42 in TMV-MP:GFP-CP was mutated to Trp by using PCR-based site-directed mutagenesis as described in Bendahmane et al. (6). The resulting clone is referred to as TMV-MP:GFP-CPT42W (Fig. 1).
Construction of pCGN-CPU1.
To construct the plasmid pCGN-CPU1, the DNA fragment DraI–KpnI (TMV nucleotides 5708–6395) containing the CP coding sequence and the 3′ nontranslated region of the viral genome was cloned into the plasmid pMON999 between EcoRI (filled in with Klenow) and KpnI. The resulting clone pMON9-CPU1 harbors an expression cassette with the CP between the 35S promoter and nos-terminator. The DNA fragment NotI-NotI from pMON9-CPU1 containing the expression cassette then was transferred to the clone pCGN1547 linearized with BamHI (23) to yield the construct pCGN-CPU1. The nucleotide sequence of all clones was confirmed by DNA sequence analysis.
In Vitro Transcription and Inoculation.
Clones that encode infectious viral RNAs were linearized with KpnI and transcribed in vitro. Transcripts were inoculated to plants as described (6). Tobacco protoplasts were prepared from BY-2 suspension cell cultures, inoculated with RNA, and transcribed in vitro via electroporation, as described (24). Infected plants and protoplasts were examined by fluorescence and laser scanning confocal microscopy, as described below.
Immunocytochemistry.
Protoplast fixation and immunostaining were performed as described (17). Polyclonal anti-CP, anti-MP, anti-replicase antibodies, and monoclonal anti-tubulin antibodies were used for immunostaining experiments. Anti-rabbit and anti-mouse antibodies conjugated with fluorescein or with rhodamine were used as secondary antibodies.
Fluorescence and Confocal Microscopy.
Infected leaf tissues and protoplasts were analyzed with a Nikon Optiphot 2-UD fluorescence microscope equipped with epi-fluorescence attachment EFD-3 and Nikon B-2A filter cube containing a 470- to 490-nm excitation filter, a DM505 nm dichroic mirror, and a BA520 barrier filter, as described (25). Confocal microscopy was conducted with an Olympus inverted microscope (IX 70) equipped with a laser scanning confocal unit. Focal planes were scanned by using a 488 Argon laser with a 550-nm barrier filter (FVX-BA550RIF) at 60× magnification (60 × 1.4 N.A. plan Apo).
Quantification of Viral Proteins.
Nontransgenic and transgenic protoplasts that produce wt CP and CPT42W were inoculated with 5 μg of wt TMV RNA produced by in vitro transcription. The relative levels of MP and CP in extracts of the protoplasts were quantitated by ELISA essentially as described in Bendahmane et al. (6). Protein extracts were used to coat the ELISA plates, and rabbit anti-MP or rabbit anti-CP antibodies were used as primary antibodies. Goat anti-rabbit antibodies conjugated with horse-radish peroxidase was used as secondary antibody; positive reactions were detected by using 2,2′-azino-bis(3-ethylbenzthiaziline-6-sulfonic acid).
Accumulation of Viral RNAs in Infected Cells.
BY-2 protoplasts were inoculated with RNA transcripts of TMV or TMV-T42W, and samples were collected from time 0 to 50 h post inoculation (h.p.i.). Total RNA was purified, and about 2 μg of RNA were analyzed by Northern blot by using standard methods (26). The DNA fragment corresponding to TMV nucleotides 5708 to 6395, which contained the CP coding and the 3′ nontranslated sequences of the viral genome, was labeled with 32P and used as probe.
Results
Distribution of CP:GFP After Infection by TMV-CP:GFP.
(i) Infection of leaf tissue.
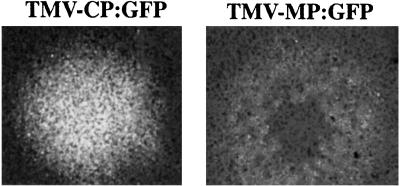
Our previous work indicated that the mutant of TMV CP referred to as CPT42W (mutation of threonine at amino acid 42 to tryptophan) gave a high level of CP-mediated resistance and limited systemic infection (6). To better understand the mechanism(s) of this effect, we initiated studies with TMV that produced fusion proteins of wt or mutant CP with GFP (TMV-CP:GFP; Fig. 1). Infectious RNA was inoculated to N. tabaccum Xanthi-nn plants. SDS/PAGE and Western blot analyses of protein extracts from infected leaves with anti-TMV CP antibody revealed the presence of the CP:GFP fusion protein of the predicted molecular mass of 44 kDa (not shown). Growing infection sites caused by TMV-CP:GFP were analyzed by fluorescence microscopy starting at one day post inoculation (dpi). At about 3 to 4 dpi, the infection sites were visible and formed very bright fluorescent discs that continued to expand over time (Fig. 2). In contrast, infection sites induced by TMV-MP:GFP resulted in a ring of fluorescence that expanded over time (Fig. 2; ref. 17).
Figure 2.
Visualization of infection sites produced by TMV-CP:GFP (fluorescent disk) on inoculated N. tabaccum Xanthi-nn through fluorescence microscopy and compared with that produced by TMV-MP:GFP (fluorescent ring).
(ii) Infection of BY-2 protoplasts.
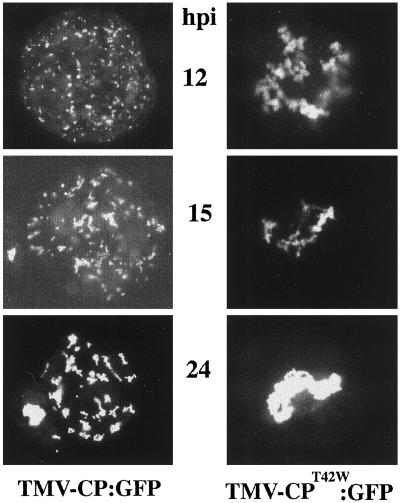
The accumulation of CP:GFP was followed by fluorescence microscopy in BY-2 protoplasts infected with TMV-CP:GFP (Fig. 3). The fluorescence of CP:GFP was first observed at about 6 h postinoculation (h.p.i.). At this time, as well as at 8 and 12 h.p.i., CP:GFP accumulated in small bodies near the periphery of the cell and around the nucleus. At 15 and 24 h.p.i., large fluorescent bodies were observed throughout the cytoplasm (Fig. 3), and were reminiscent of the virus “replication factories” described by Mas and Beachy (27) in studies of the MP:GFP fusion protein.
Figure 3.
Patterns of accumulation of CP:GFP and CPT42W:GFP in BY-2 protoplasts inoculated with TMV-CP:GFP or TMV-CPT42W:GFP, respectively. Protoplasts were inoculated with transcribed RNAs, and the accumulation of CP:GFP or CPT42W:GFP was analyzed by fluorescence microscopy. Pictures were taken at 12, 15, and 24 h.p.i.
(iii) Colocalization with other viral proteins.
To determine whether CP:GFP was colocalized with MP and replicase, the following experiment was conducted. Protoplasts were infected with TMV-CP:GFP, and samples were collected at different times after inoculation; this procedure was followed by immunocytochemistry with anti-MP, anti-126/183-kDa replicase proteins, or anti-tubulin antibodies. The CP:GFP was colocalized with the MP and with 126/183-kDa replicase proteins (data not shown), albeit at low frequency, and did not colocalize with microtubular structures. In contrast, it was shown previously that MP colocalized with microtubules in mid- and late-stages of infection and colocalized with 126/183 protein in early and middle stages of infection (27, 29).
Infection of Transgenic Plants That Express wt CP or CPT42W.
CP-MR can be overcome in plants expressing TMV CP by inoculation with viral RNA (1, 3, 4). Nontransgenic plants and transgenic plants containing wt CP (plant line 3646; ref. 1), CPT42W (plant line nn-T42W; ref. 6) or MP (plant line 277; ref. 8) were inoculated with RNA transcripts of clones TMV-CP:GFP (Fig. 1), TMV-MP:GFP (17), TMV-MP:GFP-CP (18), or TMV-ΔC-GFP (17). Infection sites were viewed by fluorescence microscopy, and those on transgenic plants were compared with infection sites on nontransgenic plants (Fig. 4).
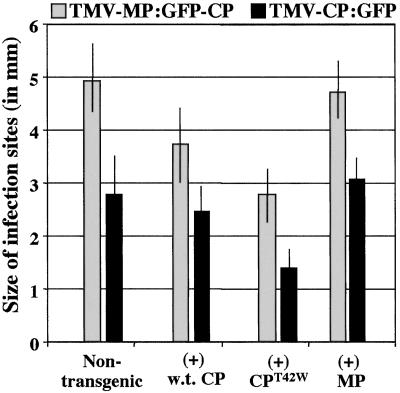
Figure 4.
Growth of infection sites on transgenic and nontransgenic tobacco plants. Growth of infection sites caused by TMV-MP:GFP-CP and TMV-CP:GFP on nontransgenic plants or transgenic plants expressing wt CP, CPT42W, or TMV MP. The diameter of infection sites (in mm) was measured at 7 days after inoculation and averaged. (Bars = SE.)
Infection of nontransgenic plants and transgenic plants containing wt CP with transcripts of TMV-MP:GFP-CP produced fluorescent rings of approximately the same size and same relative fluorescence. Similarly, infection of these plant lines with TMV-ΔC-GFP (data not shown) or TMV-CP:GFP produced fluorescent discs/spots of about the same size and relative fluorescence (Fig. 4). These results indicated that wt CP did not significantly impact the progression of the infection or production of MP:GFP (expressed from the MP subgenomic promoter) or GFP (expressed from the CP subgenomic promoter) of these viruses.
In contrast, the numbers of infection sites and the relative fluorescence of each infection site were very low on plant lines that contained CPT42W. When case infection took place, the size of the infection sites was significantly smaller on plant lines with CPT42W than on lines with wt CP or nontransgenic plants (Fig. 4). These results suggest that CPT42W interferes with accumulation or function of the MP, thereby reducing the spread of infection in transgenic plants that contain CPT42W. Furthermore, the effect was more apparent when the virus produced CP (either wt CP or CP:GFP fusion protein) than when CP was not produced by the infection. Infection of plant line 277 (containing the MP) was similar to infection of nontransgenic plants.
Infection by TMV Carrying CPT42W Fused with GFP.
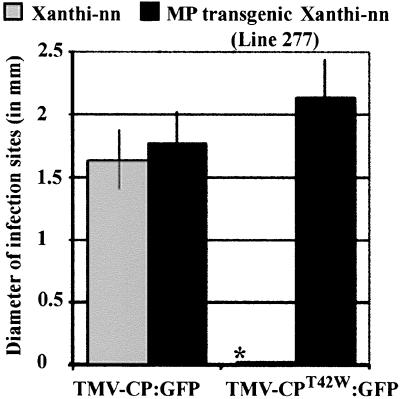
The cDNA encoding CPT42W (6) was used to replace its homologue in TMV-CP:GFP to generate the infectious cDNA clone pTMV-CPT42W:GFP (Fig. 1). RNA transcribed in vitro was inoculated to nontransgenic Xanthi-nn plants and BY-2 protoplasts. In repeated experiments with plants inoculated with TMV-CPT42W:GFP, no infection sites were obtained. When TMV-CPT42W:GFP (which encodes MP) was inoculated to transgenic tobacco plants that accumulate MP (plant line 277), infectivity was restored to wt levels (Fig. 5). Thus, when MP is present in tissues before virus infection the negative effect of CPT42W on virus infectivity is overcome.
Figure 5.
Expansion of infection sites produced by TMV-CP:GFP and TMV-CPT42W:GFP on nontransgenic N. tabaccum Xanthi-nn or transgenic plants expressing TMV MP (line 277). The diameter of infection sites (in mm) was measured at 4 dpi and averaged. For the mean size of infection sites, bars = SE. *, no infection sites detected.
In BY-2 protoplasts infected by TMV-CP:GFP, CP:GFP fusion protein accumulated in small fluorescent bodies early during infection. In contrast, infection by TMV-CPT42W:GFP resulted in accumulation of CPT42W:GFP in large fluorescent aggregates around the nucleus (Fig. 3). It was previously shown in studies of protoplasts infected with TMV-MP:GFP that MP:GFP fusion protein accumulated around the nucleus early after infection. Later in infection, MP:GFP migrated from near the nucleus to fluorescent bodies around the cytoplasm (27). In contrast, CPT42W:GFP remained around the nucleus throughout.
Taken together, these studies suggest that CPT42W restricts infection either by (i) reducing the synthesis of MP, (ii) restricting one or more of the functions of the MP produced during virus infection, or (iii) increasing the turnover of MP:GFP. To address these possibilities, we conducted experiments to compare the production and accumulation of MP and MP:GFP fusion protein during virus infection.
Accumulation of MP:GFP in the Presence of wt CP or CPT42W.
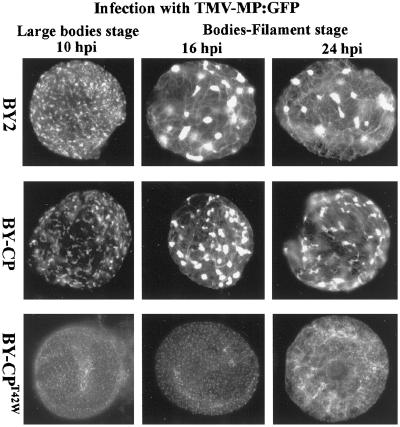
To examine further the effect of CPT42W on synthesis and accumulation of MP, the CP gene in the clone TMV-MP:GFP-CP was mutated. The resulting clone, TMV-MP:GFP-CPT42W was characterized by a very low level of fluorescence in BY-2 protoplasts and in infected plants, as predicted by previous studies with TMV-MP:GFP-CP (25). Because the low level of fluorescence restricted studies in leaves and protoplasts, we generated a transgenic line of BY-2 cells that expresses CPT42W (BY-CPT42W). Infection of BY-CPT42W protoplasts with TMV-MP:GFP was followed by fluorescence microscopy at 10, 16, and 24 h.p.i. The accumulation of MP:GFP in these protoplasts was compared with infection in nontransgenic BY-2 cells and transgenic BY-2 expressing wt CP (BY-CP; Fig. 6). In BY-CP protoplasts, the accumulation of MP:GFP was similar to that in nontransgenic BY-2 cells. Fluorescence was detected first in puncta on the surface of the protoplasts, then in small and large virus replication bodies, followed by association with microtubules and lastly, in small puncta at or near the periphery of the protoplasts (28, 29). In contrast, in transgenic BY-CPT42W protoplasts, the MP:GFP accumulated only in small puncta and, in late stages of infection, in association with short filaments (Fig. 6). The nature of these filaments is not known but are reminiscent of association of MP:GFP with microtubules (17, 28). There was little or no accumulation in virus replication bodies.
Figure 6.
Effects of CPT42W on TMV MP accumulation during virus infection. Protoplasts of nontransgenic BY2 and transgenic BY2 expressing wt CP (BY-CP) or mutant CPT42W (BY-CPT42W) were inoculated with TMV-MP:GFP RNA. The patterns and cellular distribution of MP:GFP were followed in time by using fluorescence microscopy. The structures formed by MP:GFP were compared in protoplasts from nontransgenic BY-2, BY-CP, and BY-CPT42W cells.
Transgenic wt CP and CPT42W Affect the Accumulation of the MP.
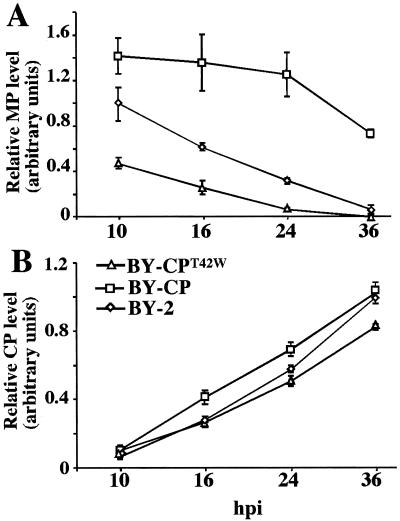
BY-2, BY-CP, and BY-CPT42W protoplasts were inoculated with wt TMV RNA, and relative amounts of MP and CP were monitored by ELISA during infection. There was a significant reduction in the level of MP accumulation in BY-CPT42W protoplasts when compared with BY-2 protoplasts (Fig. 7A). This finding demonstrates that accumulation of the MP during virus infection is negatively affected by the presence of CPT42W. In contrast, and to our surprise, the amounts of MP that accumulated in BY-CP protoplasts (expressing wt CP) were higher than in nontransgenic BY-2 and in BY-CPT42W protoplasts (Fig. 7A). This result demonstrates that wt CP enhances the accumulation of MP. In these same cells, the accumulation of CP did not differ between nontransgenic BY-2 and transgenic BY-CP protoplasts, whereas the amount of CP in BY-CPT42W protoplasts was marginally lower (Fig. 7B). These data further support the conclusion that CPT42W interferes specifically with synthesis and/or accumulation of the MP.
Figure 7.
Effects of CPT42W on TMV MP levels of accumulation during virus infection. Protoplasts of nontransgenic BY2 and transgenic BY2 expressing wt CP (BY-CP) or mutant CPT42W (BY-CPT42W) were inoculated with TMV RNA. The relative levels of accumulation of MP (A) and of CP (B) were measured and compared in protoplasts from nontransgenic BY-2, BY-CP, and BY-CPT42W cells during infection in ELISA experiments.
Infection by TMV-CPT42W Results in Lower Levels of Viral RNA than Does TMV.
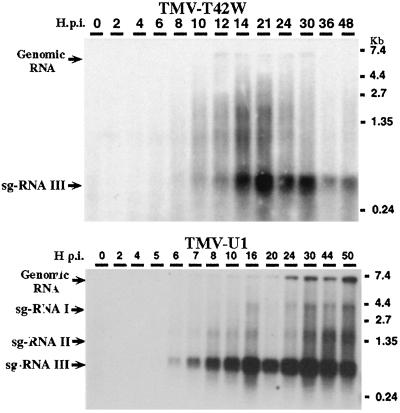
BY-2 protoplasts were inoculated with RNA transcripts TMV-T42W or wt TMV, and relative levels of viral RNAs were compared. As shown in Fig. 8, infections with TMV-T42W resulted in lower levels of all viral RNAs than did infection with wt TMV. The reduced amount of viral RNA, as well as several species of subgenomic RNAs, can be the result of reduced production or accumulation of viral RNAs. Because CP T42W is unable to encapsidate viral RNA (6), the low level of accumulation of viral RNAs may reflect reduced encapsidation plus reduced synthesis.
Figure 8.
Analyses of TMV and TMV-T42W RNA accumulation in BY-2 protoplasts. BY-2 protoplasts were inoculated with in vitro transcribed viral RNA, and samples were harvested from time 0 (time of initial contact of RNA with protoplasts) to 50 h after inoculation. Total RNA was purified and 2 μg was analyzed by Northern blot. The position of genomic RNA and subgenomic (sg) RNAs are indicated: sg-RNA I, sg-RNA II (MP subgenomic RNA), and sg-RNAIII (CP subgenomic RNA).
Discussion
We previously reported that a mutant of TMV-CP in which threonine at amino acid 42 was substituted for tryptophan (CPT42W) conferred strong CP-MR to TMV infection by inhibiting virus disassembly and by interfering with spread of infection (6). To further characterize the mechanisms of resistance conferred by CPT42W, we followed the infection of TMV-CP:GFP and TMV-MP:GFP-CP on nontransgenic plants and transgenic plants that contain wt CP, CPT42W, or wt MP. The infection sites produced by TMV-CP:GFP or TMV-MP:GFP-CP (both of which harbor the CP gene) were half the size on the nn-T42W plants containing CPT42W compared with the other plant lines. These data show that transgenic CPT42W interferes with virus cell-to-cell spread.
To investigate the role of CPT42W on replication and virus spread, TMV-CPT42W:GFP was inoculated to plants and to protoplasts. It was previously shown that TMV-CPT42W did not cause systemic infection (6). Similarly, when TMV-CPT42W:GFP was inoculated to tobacco plants, no infection sites were observed. However, infection was restored in transgenic plants that contain the MP of TMV. Together these results indicate that CPT42W interferes with the production or function of MP in cell-to-cell movement in nontransgenic plants. In BY-2 protoplasts, infection by TMV-CPT42W resulted in lower levels of MP subgenomic mRNA and viral RNA but not CP subgenomic RNA, as compared with infection by wt TMV (Fig. 8).
In another experiment, we found that in transgenic BY-CPT42W protoplasts infected with TMV RNA, the accumulation of MP was significantly lower than in nontransgenic BY-2 cells (Fig. 7A). In contrast, the accumulation of MP was greater in BY-CP protoplasts than in BY-2 protoplasts. In light of these findings, we propose that TMV CP has a specific positive regulatory function in MP production and that the mutant CP-T42W has a negative effect on MP production. However, it is not clear how CP regulates the accumulation or function of MP. Because the accumulation of CP of challenge virus did not differ between nontransgenic BY-2, transgenic BY-CP, and transgenic BY-CPT42W protoplasts, the positive effect of wt CP and the negative effect of the CP-T42W seem to be specific for the MP.
To determine how CPT42W interferes with MP function, we analyzed the infection of TMV-MP:GFP in protoplasts from transgenic BY-2 cell lines that contain CPT42W (BY-CPT42W) or wt CP (BY-CP). The accumulation of MP:GFP was very different in these protoplasts compared with nontransgenic BY-2 protoplasts. In BY-CPT42W protoplasts, MP:GFP failed to accumulate in large fluorescent bodies, which represent “replication factories” (27). Likewise, MP:GFP had reduced levels of association with microtubules in cells that contain CPT42W (Fig. 6). In contrast, patterns of accumulation of MP:GFP were similar in BY-CP and BY-2 protoplasts. These results together suggest that CPT42W interferes with the formation of virus replication complexes by interfering with the production or the function of MP.
The implications of these studies are somewhat in contrast with the results of previous work which found that mutation or deletion of CP does not notably affect TMV replication per se or cell-to-cell movement (10–12, 30, 31). Similar conclusions regarding the role of CP in virus replication were reached for other RNA-containing plant viruses, although CP may be essential for, or may facilitate, cell-to-cell spread of infection (review by Lazarowitz and Beachy, ref. 16 and refs. 32–34). We suggest that the cell system and the methods used made it possible to resolve differences in virus gene expression and in cellular aspects of infection that were not detected by other workers. In light of the results presented here, it will be informative to reexamine the effects of viral CP on production and/or function of other viral proteins.
As a result of current and prior studies, it is possible to define more completely the role of TMV CP in virus infections and in CP-mediated resistance. Both wt CP and CPT42W interfere with virus disassembly. CPT42W restricts the production or accumulation of MP, whereas wt CP has the opposite effect. We propose that reducing the amount of MP reduces the development of replication complexes and the association of MP with microtubules. One or both of these effects is likely to be the cause of reduced cell-to-cell movement in transgenic plants that express CPT42W. In light of these findings, it is clear that the immunity of plants that produce CPT42W to TMV infection is a combination of interference with virus disassembly (6) and interference with virus cell-to-cell movement by interference with the MP.
Acknowledgments
We thank Mike Dyer and Nancy Mathis for assistance with the plants. This work was supported by Grant AI27161 from the National Institutes of Health. Other support was provided by The Donald Danforth Plant Science Center and The Scripps Research Institute.
Abbreviations
- CP
coat protein
- CP-MR
CP-mediated resistance
- TMV
tobacco mosaic virus
- wt
wild type
- MP
movement protein
- GFP
green fluorescent protein
- h.p.i.
hours postinoculation
- BY-CP
BY-2 expressing wt CP
References
- 1.Powell-Abel P, Nelson R S, De B, Hoffmann N, Rogers S G, Fraley R T, Beachy R N. Science. 1986;232:738–743. doi: 10.1126/science.3457472. [DOI] [PubMed] [Google Scholar]
- 2.Beachy R N. Curr Opin Biotech. 1997;8:215–220. doi: 10.1016/s0958-1669(97)80105-x. [DOI] [PubMed] [Google Scholar]
- 3.Bendahmane M, Beachy R N. Adv Virus Res. 1999;53:369–386. doi: 10.1016/s0065-3527(08)60357-7. [DOI] [PubMed] [Google Scholar]
- 4.Register J C, III, Beachy R N. Virology. 1988;166:524–532. doi: 10.1016/0042-6822(88)90523-5. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Beachy R N, Wilson T M A, Shaw J G. Virology. 1990;179:893–895. doi: 10.1016/0042-6822(90)90163-l. [DOI] [PubMed] [Google Scholar]
- 6.Bendahmane M, Fitchen J H, Zang G H, Beachy R N. J Virol. 1997;71:7942–7950. doi: 10.1128/jvi.71.10.7942-7950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa M, Meshi T, Motoyoshi F, Takamatsu N, Okada Y. Nucleic Acids Res. 1986;14:8291–8305. doi: 10.1093/nar/14.21.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deom C M, Oliver M J, Beachy R N. Science. 1987;237:389–394. doi: 10.1126/science.237.4813.389. [DOI] [PubMed] [Google Scholar]
- 9.Meshi T, Watanabe H, Saito T, Sugimoto A, Maeda T, Okada Y. EMBO J. 1987;6:2557–2563. doi: 10.1002/j.1460-2075.1987.tb02544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel A, Hills G J, Markham R. J Mol Biol. 1966;19:140–144. doi: 10.1016/s0022-2836(66)80056-6. [DOI] [PubMed] [Google Scholar]
- 11.Atabekov J G, Dorokhov Y L. Adv Virus Res. 1984;29:313–364. doi: 10.1016/s0065-3527(08)60412-1. [DOI] [PubMed] [Google Scholar]
- 12.Dawson W O, Bubrick P, Grantham G L. Phytopathology. 1988;78:783–789. [Google Scholar]
- 13.Buck K W. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding X, Shintaku M H, Carter S A, Nelson R S. Proc Natl Acad Sci USA. 1996;93:11155–11160. doi: 10.1073/pnas.93.20.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 16.Lazarowitz S G, Beachy R N. Plant Cell. 1999;11:535–549. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinlein M, Epel B L, Padgett H S, Beachy R N. Science. 1995;270:1983–1985. doi: 10.1126/science.270.5244.1983. [DOI] [PubMed] [Google Scholar]
- 18.Oparka K J, Prior D A M, Santa Cruz S, Padgett H S, Beachy R N. Plant J. 1997;12:781–789. doi: 10.1046/j.1365-313x.1997.12040781.x. [DOI] [PubMed] [Google Scholar]
- 19.Bendahmane M, Koo M, Karrer E, Beachy R N. J Mol Biol. 1999;290:9–20. doi: 10.1006/jmbi.1999.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 21.Reichel C, Mathur J, Eckes P, Langemenkemper K, Koncz C, Schell J, Reiss B, Maas C. Proc Natl Acad Sci USA. 1996;93:5888–5893. doi: 10.1073/pnas.93.12.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt C A, Beachy R N. Virology. 1991;181:109–117. doi: 10.1016/0042-6822(91)90475-q. [DOI] [PubMed] [Google Scholar]
- 23.McBride K E, Summerfelt K R. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe Y, Meshi T, Okada Y. FEBS Lett. 1987;219:65–69. [Google Scholar]
- 25.Szécsi J, Ding X H, Lim C O, Bendahmane M, Cho M J, Nelson R S, Beachy R N. Mol Plant–Microbe Interact. 1999;12:143–152. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Mas P, Beachy R N. J Cell Biol. 1999;147:945–958. doi: 10.1083/jcb.147.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinlein M, Padgett H S, Gens J S, Picard B G, Casper S J, Epel B L, Beachy R N. Plant Cell. 1998;10:1107–1120. doi: 10.1105/tpc.10.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mas P, Beachy R N. Plant J. 1998;15:835–842. doi: 10.1046/j.1365-313X.1998.00253.x. [DOI] [PubMed] [Google Scholar]
- 30.Takamatsu N, Ishikawa M, Meshi T, Okada Y. EMBO J. 1987;6:307–312. doi: 10.1002/j.1460-2075.1987.tb04755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck K W. Phil Trans R Soc London B. 1999;354:613–327. doi: 10.1098/rstb.1999.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritzenthaller C, Schmit A-C, Michleer P, Stussi-Garaud C, Pinck L. Mol Plant–Microbe Interact. 1995;8:379–387. [Google Scholar]
- 33.Santa Cruz S, Roberts A G, Prior D A, Chapman S, Oparka K J. Plant Cell. 1998;10:495–510. doi: 10.1105/tpc.10.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackman L M, Boevink P, Cruz S S, Palukaitis P, Oparka K J. Plant Cell. 1998;10:525–538. doi: 10.1105/tpc.10.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]