Abstract
The POU homeodomain protein Oct-4 and the Forkhead Box protein FoxD3 (previously Genesis) are transcriptional regulators expressed in embryonic stem cells. Down-regulation of Oct-4 during gastrulation is essential for proper endoderm development. After gastrulation, FoxD3 is generally down-regulated during early endoderm formation, although it specifically remains expressed in the embryonic neural crest. In these studies, we have found that Oct-4 and FoxD3 can bind to identical regulatory DNA sequences. In addition, Oct-4 physically interacted with the FoxD3 DNA-binding domain. Cotransfection of Oct-4 and FoxD3 expression vectors activated the osteopontin enhancer, which is expressed in totipotent embryonic stem cells. FoxA1 and FoxA2 (previously HNF-3α and HNF-3β) are Forkhead Box transcription factors that participate in liver and lung formation from foregut endoderm. Although FoxD3 activated the FoxA1 and FoxA2 promoters, Oct-4 inhibited FoxD3 activation of the FoxA1 and FoxA2 endodermal promoters. These data indicate that Oct-4 functions as a corepressor of FoxD3 to provide embryonic lineage-specific transcriptional regulatory activity to maintain appropriate developmental timing.
Lineage commitment and differentiation of embryonic stem (ES) cells is controlled by regulatory genes that mediate permanent phenotypic change. These regulatory genes are often transcriptional regulators that activate or repress patterns of gene expression that create the phenotypic change seen during stem cell differentiation (1, 2). These transcription factors can not only mediate phenotypic maturation during a particular developmental stage but also regulate expression of the transcription factors that are important in the next stage of embryogenesis (1, 2). Although the regulation of the initial decisions in ES cell differentiation is only beginning to be understood, transcriptional regulators clearly also play important roles in cell lineage commitment and stage progression (3).
Transcriptional regulators can exhibit tissue-specific expression, and they can be sorted into related families on the basis of conserved amino acid sequences of their DNA-binding domains (4). Oct-4 and FoxD3 (previously Genesis) are two transcriptional regulators whose expression is highly limited to ES cells. Fox D3 is a member of the Forkhead Box (Fox) family (5), which has a winged-helix DNA-binding structure (6). This family is strongly implicated in early embryonic lineage decisions, especially for development of the endoderm and subsequent endodermal organogenesis (1). For example, other Fox family members, FoxA1 (previously HNF-3α) and FoxA2 (previously HNF-3β) are critical for the embryonic development of endodermal foregut organs such as the liver and lung (1).
Oct-4 is a member of the POU homeodomain family of transcriptional regulators also known to be critical in embryonic development (2). Oct-4 is expressed almost exclusively in ES cells before implantation, and is down-regulated at the blastocyst stage and gastrulation, where somatic lineages are first defined, whereas FoxD3 remains active at that stage (7). The level of Oct-4 expression drives the decision of the blastocyst to form ultimately either embryonic or extraembryonic tissue (2, 8). Specifically, high levels of Oct-4 designate cells to become extraembryonic mesoderm or endoderm such as the yolk sac, normal Oct-4 levels maintain an ES cell totipotentiality, and low levels designate cells to become trophoectoderm, such as the placenta (8). In multiple in vitro systems, Oct-4 down-regulation is essential for mammalian ES cells to differentiate to defined lineages (2, 8). Although only a few targets of Oct-4 transcriptional regulation are known (2, 8), Oct-4 can function as a homo- or heterodimer on palindromic octamer DNA sequences (classic consensus ATTTGCAT) to repress or activate transcription according to flanking sequence or chromatin structure (2). For example, Oct-4 activates expression of fibroblast growth factor-4 and osteopontin in ES cells, but represses the β-human chorionic gonadotropin promoter in ES cells (2). Oct-4 may require interacting factors such as Sox-2 or E1A to mediate transcriptional activation (2). Both Oct-4 and FoxD3 expression is down-regulated by the differentiating agent retinoic acid, which tends to drive undifferentiated cells to express neural markers (2, 5, 7, 8).
The most fundamental decision an ES cell makes is whether to remain totipotent, or to differentiate. Continued Oct-4 expression is postulated to play a critical role in maintaining the totipotent state of ES cells by maintaining activation of appropriate primitive genes, and by repressing more differentiated lineage-specific promoters (2, 8). However, few such lineage-specific targets are known, and the mechanism by which Oct-4 might mediate such regulation is not known.
In this study, we found that FoxD3 and Oct-4 both bound to and transcriptionally activated the osteopontin enhancer, which is expressed in ES cells. However, although FoxD3 bound to and activated the promoters of the early endodermal transcription factors FoxA1 and FoxA2, Oct-4 completely inhibited this activation by physically interacting with the DNA-binding domain of FoxD3. However, Oct-4 on its own did not bind to or activate the FoxA1 or FoxA2 promoters. Thus, Oct-4 can prevent ES cell lineage differentiation gene expression cascades by functioning as a corepressor of lineage-specific transcription factors such as FoxD3.
Materials and Methods
Reporter Assays.
All transfections were performed in the human embryonic kidney 293 cell line by using calcium phosphate as described (9). Transfection efficiencies were normalized with a Rous sarcoma virus-β-galactosidase expression vector. DNA concentrations transfected were normalized by using empty expression vectors. Cells were harvested 48 h after transfection, and luciferase activity was measured by using Galacto-Light Plus according to the manufacturer's instructions (Amersham Pharmacia). Chloramphenicol acetyltransferase (CAT) activities were measured by using [14C]chloramphenicol and acetyl-CoA as we described (10). The expression vectors for PJ6-Fox D3, pCMV-Oct-4, pCMV-HNF3α (here FoxA1), and pCMV-HNF3β (here FoxA2) have been described (5, 11–13). The reporter constructs 6PORE-luciferase, HNF-3α-CAT (here designated FoxA1-CAT), and HNF3β-CAT (here FoxA2) have also been described (11–13). Winged-helix/Forkhead gene nomenclature is described at www.biology.pomona.edu/fox.html.
Recombinant Protein Production.
The Forkhead box DNA-binding domains of FoxA1, FoxA2, and FoxD3 were cloned into the expression vector pGEX2T, and glutathione S-transferase (GST) fusion protein was expressed in BL21(DE3)plysS Escherichia coli and purified to homogeneity on a glutathione-Sepharose column as we described (14). The Oct-4 cDNA was cloned into pET24-TEV-His-6 expression vector, expressed in BL21(DE3)plysS E. coli, and purified to homogeneity on a Ni-NTA agarose column as we described (11).
Electrophoretic Mobility-Shift Assays (EMSAs).
The duplex oligonucleotide probes used were as follows: 6PORE, 5′-AAGTTAAAATCACATTTGAAATGCAAATGG (osteopontin enhancer sequence; refs. 11 and 15); FoxA1, 5′-TTACACTGCTTTGTAAACAAAGTGAGGG (Forkhead box in the FoxA1/HNF-3α promoter; ref. 12); and FoxA2, 5′-CACCTACTGCCCTGTTTGTTTAGTTACG (Forkhead box in the FoxA2/HNF-3β promoter; ref. 13). Purified protein was incubated in 25 μl of binding buffer [10 mM Hepes, pH 7.9/60 mM KCl/1 mM EDTA/7% glycerol/0.25 mg/ml BSA/100 μg/ml poly(dI-dC)/8 mM DTT/4% FCS] with or without unlabeled competing duplex oligomer probe on ice for 15 min. The sample was then incubated with 104 cpm of 32P-labeled probe at room temperature for another 15 min. Samples were then electrophoresed on a prerun 5% nondenaturing polyacrylamide gel (acrylamide/bisacrylamide, 29:1) in 0.5× TBE buffer (1× = 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 10 V/cm. The gel was dried and autoradiographed at −70°C overnight.
Immunoprecipitation.
For the GST pull-down experiments, 10 ng of GST-FoxD3 fusion protein was incubated with 10 ng of Oct-4-His protein for 20 min at room temperature in 100 μl of Tris-buffered saline. The sample was then mixed with glutathione-Sepharose for 20 min at room temperature. The sample was washed three times in Tris-buffered saline and boiled in 1× protein-loading buffer for 5 min. Denatured samples were run on an SDS/polyacrylamide gel at 200 V for 30 min. Protein was transferred to a poly(vinylidene difluoride) membrane and blocked with 5% nonfat milk for 1 h. For the GST in vitro pull-down, Oct-4 protein was detected by 1:2,500 dilution of anti-Oct-4 antibody by using chemiluminescence (Amersham Pharmacia) as we described (16). The GST pull-down membranes were stripped by boiling in 1× SSC/0.1% SDS/200 mM Tris, pH 6.8, and reprobed with anti-GST at a 1:5,000 dilution as a control. For the in vivo immunoprecipitation, 10 μg of the FoxD3 expression vector and 10 μg of the Oct-4 expression vector were cotransfected as above into 293 cells, cell lysates obtained after 48 h, and incubated on ice with 1:500 Oct-4 antisera for 30 min. After adding protein G beads and gently rocking for 30 min, the sample was microcentrifuged, and the beads were washed three times with Tris-buffered saline. After boiling in 1× protein-loading buffer, the sample was electrophoresed as above on an SDS/polyacrylamide gel, and transferred as above to a poly(vinylidene difluoride) membrane. The presence of FoxD3 immunoprecipitated with Oct-4 was detected by 1:1,000 dilution of FoxD3 antibody by using chemiluminescence (16).
Results
Sequence-Specific DNA Binding of Oct-4 and FoxD3.
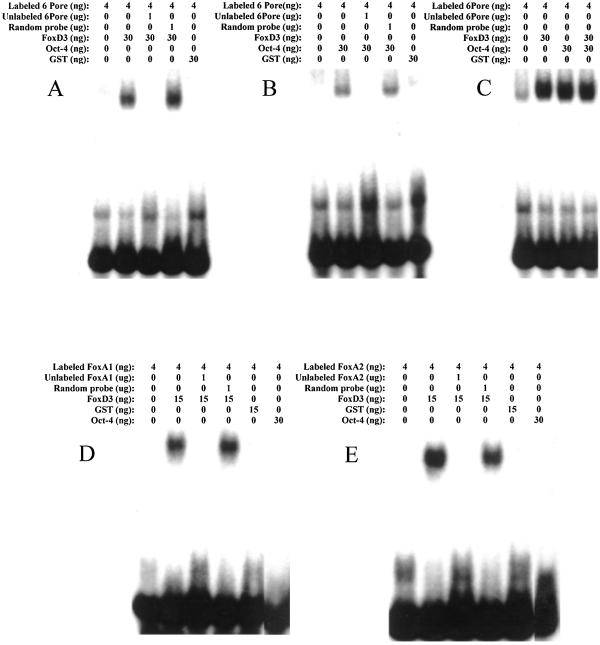
On the basis of previous studies, we had noted that the nucleotide-binding sequences of FoxD3 and Oct-4 were similar (5, 11, 15). Therefore, EMSA was used to study the DNA-binding specificity of purified recombinant FoxD3 and Oct-4 protein. Radiolabeled duplex oligonucleotides were incubated with either Oct-4 or FoxD3 alone or together, and then electrophoresed on nondenaturing polyacrylamide gels to visualize the slowly migrating protein–DNA complexes. In Fig. 1 A and B, EMSA shows the decreased mobility of the osteopontin enhancer duplex radiolabeled oligonucleotide (6PORE) when bound either FoxD3 or Oct-4. These protein–DNA complexes were shown to be specific by competition with unlabeled 6PORE but not with random duplex oligonucleotide. When FoxD3 and Oct-4 were both incubated together with labeled 6PORE, the mobility of the protein-DNA complex did not decrease, indicating that they bound to the DNA sequence independently of interacting with each other (Fig. 1C). This independent action also shows that it is unlikely that each protein bound 6PORE at the same time.
Figure 1.
EMSAs of the binding of recombinant FoxD3 and Oct-4 to the osteopontin enhancer 6PORE sequence (A–C) and the Forkhead Boxes in the FoxA1 and FoxA2 (D, E) promoters. Specific unlabeled duplex oligonucleotides competed for the DNA binding, as indicated by a loss of the mobility shift, whereas random unlabeled duplex oligonucleotides had no effect on the DNA binding. The concentration of proteins used was chosen on the basis of previous titrations, using the lowest concentration that gave the maximum mobility shift. The Forkhead proteins bound DNA more efficiently than Oct-4, most likely because Oct-4 needs to dimerize before DNA binding.
However, although FoxD3 was able to bind to the Forkhead box DNA sequences from the FoxA1 and A2 promoters, Oct-4 was unable to bind to those sequences. Fig. 1 D and E shows that the labeled FoxA1 and FoxA2 Forkhead Box duplex oligonucleotides exhibited specific mobility shifts when in the presence of FoxD3. Recombinant purified FoxA1 and FoxA2 proteins also bound to the cognate Forkhead Box sequence within their respective promoters. However, no supershifts occurred when FoxA1 or FoxA2 protein was incubated with FoxD3, indicating that these proteins bound DNA independently (data not shown).
These data show that although both Oct-4 and FoxD3 specifically bound the osteopontin enhancer, only FoxD3 was able to bind to the endodermal FoxA1 and FoxA2 promoters.
Transcriptional Regulation of Embryonic Promoters by Oct-4 and FoxD3.
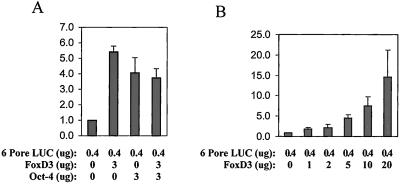
To assess whether FoxD3 could regulate transcription through the Oct-4 target, the osteopontin enhancer, cotransfection assays were performed. A FoxD3 expression vector was transfected with 6PORE-luciferase, an osteopontin reporter plasmid, into 293 embryonic kidney cells. Fig. 2 A and B shows that FoxD3 activates transcription through the osteopontin enhancer in a dose-dependent manner. Oct-4 also activates expression of the 6PORE reporter, either alone or in combination FoxD3.
Figure 2.
Cotransfection assays of Oct-4 and FoxD3 expression vectors with the 6PORE-luciferase reporter. This 6PORE reporter vector contains sequences from the osteopontin enhancer (15). The y axis represents relative luciferase units. (A) Both Oct-4 and FoxD3 activated transcription from this reporter, but did not increase transcription when transfected together. (B) FoxD3 displayed a dose-dependent activation of the 6PORE reporter.
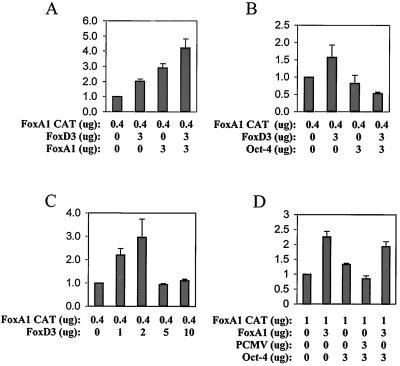
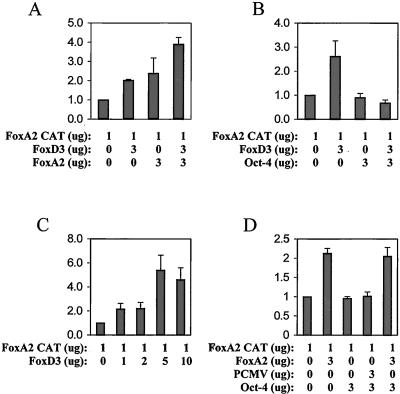
Cotransfection experiments were performed to assess whether FoxD3 could also activate transcription through the FoxA1 and FoxA2 endodermal promoters. Figs. 3 and 4 show that FoxD3 activates transcription through both the FoxA1 and FoxA2 promoters in a dose-dependent manner. However, at higher levels of FoxD3, a loss of this activation occurred, indicating that such activation depends on appropriate intracellular levels of FoxD3 (Figs. 3C and 4C). Although FoxA1 and FoxA2 activated their own promoters, no synergism with FoxD3 occurred, only additive transcriptional activation. However, Oct-4 alone did not activate either of the FoxA1 or FoxA2 promoters, consistent with the EMSA finding that Oct-4 did not bind to the FoxA1 or FoxA2 promoter Forkhead Box sequences. Significantly, Oct-4 blocked the activation of either promoter by FoxD3 (Figs. 3 and 4). Thus, although Oct-4 could not bind (Fig. 1) the FoxA1 or FoxA2 promoters, it could repress FoxD3 activation of those promoters. However, Oct-4 did not repress the activation by FoxA1 or FoxA2 of their own promoters, as it did with FoxD3 (Figs. 3D and 4D).
Figure 3.
Cotransfection assays of Oct-4, FoxD3, and FoxA1 expression vectors with the FoxA1 promoter reporter. The y axis represents relative luciferase units. (A) Both FoxA1 and FoxD3 activate the FoxA1 promoter, but the activation is additive, not synergistic. (B) Oct-4 inhibits almost completely the activation of the FoxA1 promoter by FoxD3. (C) FoxD3 displayed a dose-dependent activation of the FoxA1 promoter at lower levels, but at higher levels failed to activate the promoter. (D) Oct-4 alone does not inhibit FoxA1 activation of its own promoter.
Figure 4.
Cotransfection assays of Oct-4, FoxD3, and FoxA2 expression vectors with the FoxA2 promoter reporter. The y axis represents relative luciferase units. (A) Both FoxA2 and FoxD3 activate the FoxA1 promoter, but the activation is additive, not synergistic. (B) Oct-4 inhibits almost completely the activation of the FoxA2 promoter by FoxD3. (C) FoxD3 displayed a dose-dependent activation of the FoxA2 promoter at all concentrations tested. (D) Oct-4 alone does not inhibit FoxA2 activation of its own promoter.
Physical Association of Oct-4 with FoxD3.
Recombinant purified GST protein alone or GST-FoxD3 protein (DNA-binding domain) was incubated with recombinant purified Oct-4 protein, and then the GST proteins were isolated by using glutathione-Sepharose. Western blot analysis was then performed by using antisera against Oct 4 (Fig. 5). High levels of Oct-4 were detected from the GST-FoxD3 reaction, but not in the GST-alone reaction, indicating that the DNA-binding domain of FoxD3 physically interacted with Oct-4 protein. Intracellular immunoprecipitation experiments with FoxD3 and Oct-4 cotransfected 293 cells were also performed (Fig. 5). After immunoprecipitation with anti-Oct-4 antibody, Western blot analysis was then performed with antisera against FoxD3. FoxD3 protein was also present in the cellular immunoprecipitation of Oct-4, which showed that Oct-4/FoxD3 interactions could be detected in vivo.
Figure 5.
GST pull-down assay of the interaction of FoxD3 with Oct-4. Recombinant purified GST-FoxD3 protein but not GST alone could pull down abundant Oct-4 protein, as assessed by Western blot analysis. In addition, after cotansfection of Oct-4 and FoxD3 expression vectors into 293 cells, immunoprecipitation with anti-Oct-4 and subsequent Western blot analysis with anti-FoxD3 indicated that this interaction also took place intracellularly.
Discussion
Although both Oct-4 and FoxD3 are highly expressed in ES cells, Oct-4 is down-regulated at gastrulation, an earlier stage of development than FoxD3 (17). Oct-4 is essential for maintaining the undifferentiated state of ES cells and may play a role in the malignant equivalent of ES cells, testicular or ovarian germ cell tumors (2, 8, 15). Similar to Oct-4, FoxD3 bound to and activated transcription from the 6PORE osteopontin enhancer sequence. Osteopontin is usually expressed early in the blastocyst stage, and is thought to be important for blastocyst implantation in the uterine wall (2, 8, 11, 15, 17). When both are present, no further activation of this enhancer occurs beyond either Oct-4 or FoxD3 alone. Consistent with this finding, the mobility-shift data showed that no supershift of the 6PORE probe takes place when Oct-4 and FoxD3 are present together, which implies that Oct-4 and FoxD3 bind to the same sequence independently of each other.
FoxD3 both bound to and activated transcription from the FoxA1 and FoxA2 promoters. The biphasic response of the FoxA1 promoter to FoxD3 activation, where at high concentrations of FoxD3 the promoter is activated less, could imply a negative feedback loop where another target of FoxD3 activation inhibits FoxD3. Such a negative feedback loop is common among transcription factors. Alternatively, high levels of FoxD3 could produce a locally inhibitory transcriptional effect at the promoter itself by recruiting nuclear corepressors when certain concentrations are achieved to overcome low affinities.
More significantly, Oct-4 neither bound to the Forkhead Box in the FoxA1 or FoxA2 promoters that FoxD3 bound to, nor did it activate transcription from these promoters. Significantly, Oct-4 blocked the transcriptional activation of the FoxA1 and FoxA2 promoters by FoxD3. This repression was specific for FoxD3. Oct-4 did not repress the activation of the FoxA1 or FoxA2 promoters by FoxA1 or FoxA2 proteins. Immunoprecipitation studies found that Oct-4 could physically interact with the DNA-binding domain of FoxD3, which implies that when Oct-4 is not binding to DNA it can function as a corepressor to inhibit the lineage-specific promoters used here. It is possible that the dimerization and conformational changes that Oct-4 undergoes when it binds DNA prevent it from acting as a corepressor of FoxD3. When Oxt-4 is not binding DNA, then it can bind to the FoxD3 DNA-binding domain and repress FoxD3 transcriptional activation.
Two possible mechanisms for the inhibition of FoxD3 activation of FoxA1 or FoxA2 exist. First, Oct-4 could inhibit FoxD3 activation of the FoxA1 or FoxA2 promoters by blocking binding of FoxD3 to the Forkhead Box sequence in those promoters. However, the 6PORE mobility shift assays did not show any change in binding when both Oct-4 and FoxD3 are present together. Second, Oct-4 could function as a true corepressor by decreasing FoxD3 interaction with the transcriptosome apparatus. In either case, this inhibition prevents inappropriate activation of endodermal promoters in a totipotent ES cell. When Oct-4 is down-regulated after gastrulation and the initial formation of the primitive endoderm, then FoxA1 and FoxA2 can be activated appropriately by the FoxD3, which is still present. Once activated, the proteins these promoters generate will maintain expression throughout organogenesis (Figs. 3 and 4), even as FoxD3 is down-regulated.
In previous studies with adult cells (where FoxD3 is not usually expressed) as opposed to embryonic cells, FoxD3 repressed transcription from a reporter plasmid containing a truncated thymidine kinase promoter and its consensus DNA-binding site (5). That study and the data here imply that FoxD3 modifies its transcriptional regulatory activities on the basis of the cellular and promoter context. Thus, either corepressors in adult cells or coactivators in embryonic cells likely modify FoxD3 activity, or FoxD3 has specific activity based on the DNA sequence it binds. The data presented here show that Oct-4 modifies its activity on the basis of the promoter context, and whether it is bound to DNA, not just the cellular context.
The finding that Oct-4 can serve as a corepressor to prevent activation of genes that would drive the ES cell to differentiation is consistent with the current hypothesis of Oct-4 function (2, 8, 11). The intriguing model that best explains these data is that a set of transcriptional activators are present in the ES cell, such as FoxD3, ready to initiate early lineage-specific gene expression, but they are prevented from doing so until Oct-4 is down-regulated around gastrulation. If Oct-4 maintains a proliferative, undifferentiated ES cell state, it is a potential therapeutic target for the treatment of germ cell neoplasia.
Acknowledgments
K.R. is supported by the Lance Armstrong Foundation. R.H. is supported by National Institutes of Health Grants PO1 CA74295 and HL48914. R.H.C. is supported by National Institutes of Health Grant GM43241. H.S. is supported by the Marion Dilley and David George Jones Funds, and the Commonwealth and General Assembly of Pennsylvania.
Abbreviations
- ES
embryonic stem
- GST
glutathione S-transferase
- EMSA
electrophoretic mobility-shift assay
References
- 1.Hromas R, Costa R. Crit Rev Oncol Hematol. 1995;20:129–140. doi: 10.1016/1040-8428(94)00151-i. [DOI] [PubMed] [Google Scholar]
- 2.Pesce M, Scholer H R. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 3.Reid L. Cell. 1990;63:875–882. doi: 10.1016/0092-8674(90)90491-v. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell P J, Tjian R. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 5.Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada D, Fletcher C, Jenkins N, Copeland N, Klemsz M, Hromas R. J Biol Chem. 1996;271:23126–23133. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- 6.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 7.Hromas R, Ye H, Spinella M, Dmitrovsky E, Xu D, Costa R H. Cell Tissue Res. 1999;297:371–382. doi: 10.1007/s004410051365. [DOI] [PubMed] [Google Scholar]
- 8.Niwa H, Miyazaki J, Smith A G. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 9.Nakshatri H, Chambon P. J Biol Chem. 1994;269:890–902. [PubMed] [Google Scholar]
- 10.Klemsz M J, Maki R A, Papayannopoulou T, Moore J, Hromas R. J Biol Chem. 1993;268:5769–5773. [PubMed] [Google Scholar]
- 11.Tomilin A, Remenyi A, Lins K, Bak H, Leidel S, Vriend G, Wilmanns M, Scholer H R. Cell. 2000;103:853–864. doi: 10.1016/s0092-8674(00)00189-6. [DOI] [PubMed] [Google Scholar]
- 12.Peterson R S, Clevidence D E, Ye H, Costa R H. Cell Growth Differ. 1997;8:69–82. [PubMed] [Google Scholar]
- 13.Pani L, Quian X B, Clevidence D, Costa R H. Mol Cell Biol. 1992;12:552–562. doi: 10.1128/mcb.12.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overdier D G, Porcella A, Costa R H. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross M K, Vriend G, Scholer H R. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christopherson K, Campbell J, Hromas R. Blood. 2001;98:3562–3568. doi: 10.1182/blood.v98.13.3562. [DOI] [PubMed] [Google Scholar]
- 17.Furhmann G, Chung A C, Jackson K J, Hummelke G, Baniahmad A, Sutter J, Sylvester I, Scholer H R, Cooney A J. Dev Cell. 2001;1:377–387. doi: 10.1016/s1534-5807(01)00038-7. [DOI] [PubMed] [Google Scholar]