Abstract
Often as an epidemic spreads, the leading front is irregular, reflecting spatial variation in local transmission rates. We developed a methodology for quantifying spatial variation in rates of disease spread across heterogeneous landscapes. Based on data for epidemic raccoon rabies in Connecticut, we developed a stochastic spatial model of rabies spread through the state's 169 townships. We quantified spatial variation in transmission rates associated with human demography and key habitat features. We found that large rivers act as semipermeable barriers, leading to a 7-fold reduction in the local rates of propagation. By combining the spatial distribution of major rivers with long-distance dispersal we were able to account for the observed irregular pattern of disease spread across the state without recourse to direct assessment of host-pathogen populations.
Understanding the spatial dynamics of an infectious disease is critical in any attempt at predicting its emergence or spread to new geographic regions. Often as a disease spreads, the leading front of the epidemic is jagged and irregular, reflecting spatial variation in the distribution of susceptible hosts, variation in local contact rates, rare long-distance contagion, or the process of surveillance (1). Unfortunately, information on those factors influencing spatial variation in transmission rates between host and pathogen populations often is not readily available and can be very expensive to obtain, especially for those diseases primarily associated with wildlife (2). Moreover, available data primarily reflect politically defined reporting units rather than biologically relevant ecological units. Given these limitations in the structure of available data, we explored a different methodology for uncovering the basic ecological processes of disease spread across environmentally heterogeneous landscapes.
The epidemic spread of rabies has proven to be an extremely useful system for exploring a variety of approaches to spatial disease dynamics. Modeling efforts have focused primarily on the post-World War II emergence and spread of rabies in red foxes across Western Europe. The modeling approach most widely adopted employs systems of partial differential equations corresponding to reaction-diffusion processes. This elegant approach was first applied to rabies by Murray et al. (3); extensions of the theory have been reviewed critically by Mollison (4). The reaction-diffusion approach has been extended also to incorporate environmental and habitat heterogeneity (5). However, the reaction-diffusion approach has largely been directed at constructing hypothetical epidemics that fit the global features of rabies spread such as the average velocity of the wavefront rather than utilizing large databases from surveillance networks to capture the details of the spatial pattern of spread. In this paper we take an explicitly data-based approach. By using an extant database on the temporal-spatial occurrence of epidemic raccoon rabies in Connecticut, we developed a probabilistic and environmentally heterogeneous spatial model for rabies spread through the state's 169 townships.
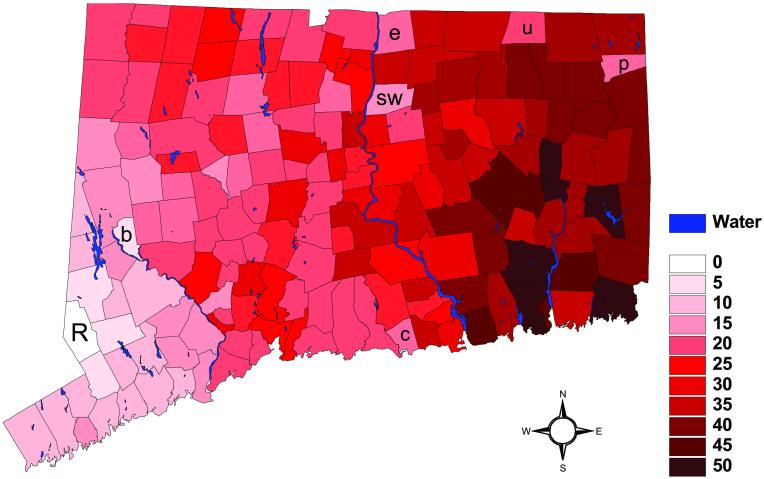
Epidemic rabies among raccoons in Connecticut first appeared in the western townships bordering New York in 1991 (6, 7). The epidemic then spread as an irregular wave across the state over a 5-year period (Fig. 1). Expansion of raccoon rabies through Connecticut was the extension of a broad front of an epidemic originating along the Virginia/West Virginia border in the mid-1970s (6, 8, 9) involving a variant of rabies virus highly adapted to raccoons (10, 11). The State of Connecticut Department of Health maintains a descriptive database consisting of monthly reports of the number of animal rabies cases in each of the 169 townships (6). Rates of local propagation of rabies could be influenced by variation in raccoon population densities and dispersal patterns associated with spatially heterogeneous habitats.
Figure 1.
The month when rabies was first observed in each township, Oi, relative to the month of the first reported case in Ridgefield Township (March, 1991) in western Connecticut. The letters are the first names of townships discussed in the text: Ridgefield (R), Enfield (e), Union (u), Putnam (p), Clinton (c), Bridgewater (b), and South Windsor (sw).
Direct assessment of raccoon populations at the spatial scale of a state would not be practical or possible. As a surrogate for assessing raccoon population densities directly, we used different variables known to be associated with raccoon population densities or suspected as determinants of the dispersal rate of rabies through a susceptible population.
Methods
We looked for associations between the rate of spread and human population densities, because raccoon population density can be influenced by the animals' association with humans, and some of the highest raccoon densities have been found in suburban or urban park settings (12, 13). Furthermore, each reported case of rabies is an interaction between a human and a rabid raccoon, thus human population density is an integral part of the detection process. Also, physiographic features such as rivers and mountains have been implicated as barriers to raccoon dispersal and potential impediments to epidemic rabies spread (14, 15). However, none of these effects have been quantified nor have any of these ecologically relevant variables been incorporated into predictive models of rabies spatial dynamics.
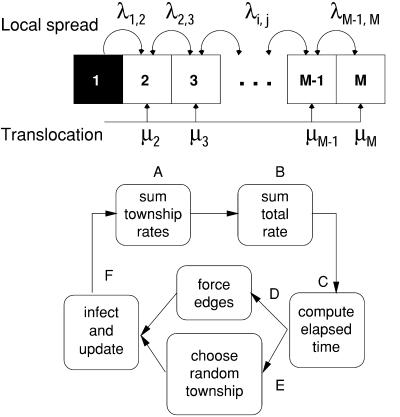
To explore the spatial dynamics of this disease, we constructed an interaction network based on the adjacency of townships (Fig. 2); two townships were considered to be adjacent if they shared at least one point along the border in common. In the simulations, the rate of disease spread into a target township depended on the fraction of townships adjacent to the target that were experiencing rabies among raccoons already, which we hereafter refer to as “infected.” The local rates of transmission across adjacent townships, λi,j, varied to incorporate heterogeneous variables; if the ith and jth townships are not adjacent, then λi,j = 0. Long-distance translocation was modeled as a low, constant rate of infection for all uninfected townships, μj, regardless of whether they had infected neighbors.
Figure 2.
A stochastic model was used to simulate the heterogeneous spread of raccoon rabies on an irregular network, illustrated here on a simple array. An infected township, i, infects its adjacent neighbor, j, at a rate λi,j. In addition, a township, j, may become infected because of translocation of rabid raccoons at a rate μj. Heterogeneity was incorporated by allowing the local rates from the neighbors, {λi,j}, and the rate of translocation, {μj}, to be different, possibly unique in different models. Each algorithm for associating a set of rates with rivers or human population density defines a stochastic candidate model. The simulation algorithm involved six steps. (A) For each township, add the rates from all possible routes of infection. (B) Add the township rates to compute a total rate. (C) Draw a random number to determine the elapsed time. (D) Check to see if any of the edges had become infected in the elapsed interval. (E) If no edges were forced, select a random township to infect. (F) Infect the forced edge or the infected township, update the local rates, and repeat until each township becomes infected.
Spatial heterogeneity was incorporated into this interactive network model by allowing the local rates of spread, the rate of long-distance translocation, to vary among the townships depending on their human demographic or geographic features. We compared five specific models incorporating different combinations of human demographic and geographic spatial heterogeneity (Table 1). The simplest model (Null) did not include any heterogeneity; spatial spread was homogeneous. In slightly more complicated models, the rate of spread was correlated linearly with the log of human population density (Human) or was lower when two townships were separated by a river and higher when they were not (River). In the most complicated heterogeneous model, the rates of local spread were linear functions of the log of human population density with different slopes and intercepts for pairs of adjacent townships depending on whether they were separated by a river (RivHum1).
Table 1.
The five candidate models used to predict the spatial dynamics of rabies epidemics on heterogeneous landscapes
| Model | Local no river (↔) river (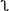 ) ) |
Translocation | χ2 (P value) df = 158 − k | χ2 (P value) df = 157 − k | k | Fitted parameters |
|---|---|---|---|---|---|---|
| Null | α | μ | 225 (2 × 10−4) | 209 (0.004) | 2 | α = 0.47 μ = 3 × 10−4 |
| Human | α + γHi | μ | 222 (3 × 10−4) | 205 (0.007) | 3 | α = 0.42, γ = 0.02 μ = 2 × 10−4 |
| River | α (↔) β (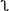 ) ) |
μ | 198 (0.01) | 183 (0.06) | 3 | α = 0.66, β = 0.09 μ = 2 × 10−4 |
| RivHum1 | α + γHi (↔) β + ρHi (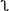 ) ) |
μ | 197 (0.01) | 182 (0.05) | 5 | α = 0.68, γ = 0.002 β = 0.05, ρ = 0.004 μ = 2 × 10−4 |
| RivHum2 | α (↔) β (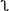 ) ) |
μ + γHi | 197 (0.01) | 182 (0.06) | 4 | α = 0.68, μ = 2 × 10−4 β = 0.1, γ = 7 × 10−6 |
Each model used a different algorithm to relate local and global rates of spread to the variables that are surrogates for raccoon habitat. In some models, the rate was a linear combination of the natural log of human population density, Hi (median, 6; range, 3–9). In other models, the rate was different if the two centroids were separated by a river or other body of water (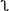 ) than if they were not (↔). Each model was fit by finding the parameters that minimized the χ2 statistic, ∑i(Oi − Ei)2/Ei. For each model, the best fit was computed with and without Putnam, a clear outlier (reported parameters reflect the fitting without Putnam). Because the 10 townships on the western border were forced, they were not included in the sum. The P values were generated from the χ2 statistic with the degrees of freedom (df) either 158 or 157 less the number of parameters in the model algorithm (k). For example, RivHum1 and RivHum2 both have a χ2 of 182, but the P values are different, because RivHum1 uses one more parameter.
) than if they were not (↔). Each model was fit by finding the parameters that minimized the χ2 statistic, ∑i(Oi − Ei)2/Ei. For each model, the best fit was computed with and without Putnam, a clear outlier (reported parameters reflect the fitting without Putnam). Because the 10 townships on the western border were forced, they were not included in the sum. The P values were generated from the χ2 statistic with the degrees of freedom (df) either 158 or 157 less the number of parameters in the model algorithm (k). For example, RivHum1 and RivHum2 both have a χ2 of 182, but the P values are different, because RivHum1 uses one more parameter.
We assumed that nonlocal rabies spread occurred as a constant background rate of infection, corresponding to the possibility of long-distance translocation of rabid raccoons by humans. We modeled global transmission as a constant rate rather than one that changed dynamically, because translocation might occur from anywhere including areas outside Connecticut that we did not model. Translocation has been known to occur by a variety of mechanisms including deliberate transport for restocking hunting areas and accidental transport by garbage trucks (6, 16). In the first four models (Null, Human, River, and RivHum1) the rate of translocation was the same everywhere, but in the fifth model (RivHum2), we let the rate of long-distance translocation be associated with the log of human population density.
The basic algorithm we used to simulate an epidemic involved six steps. First, we computed the total rate of infection in the jth township, ρj, where ρj = μjXj + ∑iλi,jXj(1 − Xi), Xj = 1 if the jth township is uninfected, and Xj = 0 otherwise. Second, we computed the total rate of infection for all townships, Λ = ∑jρj. Third, we computed the waiting time before a township becomes infected next; we assume waiting times are distributed exponentially with rate parameter Λ. Fourth, the townships on the western border of Connecticut were forced. By forced we mean that infection in these townships was not simulated; once the time index of the simulation passed the time when rabies was observed, their status was changed from uninfected to infected. These townships were part of the advancing epidemic front moving from the west, and thus we used the date when rabies was first observed in raccoons to establish boundary conditions for the advancing wave into Connecticut. Fifth, if no townships were forced, a random township was chosen; townships that had higher rates of infection had higher probabilities of becoming infected. Random townships were chosen from the multinomial distribution; the probability that the ith township was chosen was ρi/Λ. Finally, the state of the infected township was updated, and the algorithm was iterated until all townships were infected. The data and computer code are available at http://medschool.umaryland.edu/departments/epidemiology/dsmith/rabies.html.
We fit each model to the data using a stochastic global optimization procedure. We let Oi denote the observed time of first appearance of rabies in the ith township. The set of observations, Oi, constitutes the raw data against which we could evaluate the predictive power of alternative epidemic models. For a particular set of parameters, we simulated the epidemic 5,000 times to generate an expected date of first rabies appearance, Ei, for each township. For each model, we searched parameter space to find the parameters that minimized the χ2 statistic, Y = ∑i(Oi − Ei)2/Ei. Because the value returned by the simulator with 5,000 repetitions is variable, the goodness of fit is an approximation. We verified that the parameters were approximately local maxima by checking the goodness of fit measure in a neighborhood around the best-fit parameters.
In addition to comparing the goodness of fit of the models to one another, we used the χ2 distribution (with appropriate degrees of freedom) to test the overall goodness of fit of the models to the data. For each township and each model, we have used the fitted parameters to generate an expected date of appearance at each township with residuals, ɛi = Oi − Ei. We know nothing a priori about the distribution of the residuals, {ɛi}, or the elements of the χ2 statistic, {ɛ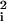 /Ei}. A histogram of the residuals shows that they are approximately normally distributed. Moreover, the elements of the χ2 statistic have a mean of 1.1 and variance of 3.6, consistent with a χ2(1) which has a mean of 1 and variance of 2. Three townships, South Windsor, Union, and Enfield, contribute disproportionately to the variance; without these three points the mean would be 0.98, and the variance would be 1.74. Because the assumptions are consistent with the χ2(1) distribution, we used the χ2 with the appropriate degrees of freedom to generate P values to test for the goodness of fit. (additional information is available at http://medschool.umaryland.edu/departments/epidemiology/dsmith/rabies.html)
/Ei}. A histogram of the residuals shows that they are approximately normally distributed. Moreover, the elements of the χ2 statistic have a mean of 1.1 and variance of 3.6, consistent with a χ2(1) which has a mean of 1 and variance of 2. Three townships, South Windsor, Union, and Enfield, contribute disproportionately to the variance; without these three points the mean would be 0.98, and the variance would be 1.74. Because the assumptions are consistent with the χ2(1) distribution, we used the χ2 with the appropriate degrees of freedom to generate P values to test for the goodness of fit. (additional information is available at http://medschool.umaryland.edu/departments/epidemiology/dsmith/rabies.html)
Results
In the models that generated the best fit (Table 1), a slower local spread of rabies was strongly associated with river crossings, the global spread by translocation was relatively frequent, and human population density had very little effect on the local spread of rabies. All the models that incorporated slowing at rivers had a better fit than the alternative models that did not include this factor (Table 1). Using the full data set, all the models differed significantly from the observed data. The model predictions and observations were substantially different in a few townships (Fig. 3 A and B).
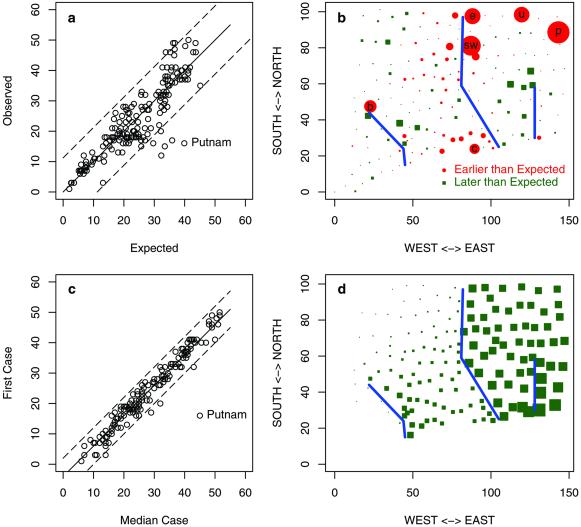
Figure 3.
The data and output from the fitted model River. (a) The expected vs. observed date of first appearance. (b) The elements of the χ2 statistic, (Oi − Ei)2/Ei, plotted at the centroid of each township. In red circles the first observed case of rabies was earlier than that predicted by the model; outliers are Putnam (p), South Windsor (p), Union (u), Clinton (c), Enfield (e), and Bridgewater (b). In green squares, the first observed case was later than the prediction. (c) The date of the first appearance vs. the date of the median case; by this criterion, Putnam clearly is an outlier. (d) The epidemic simulated without rivers and plotted by the associated delay. According to the models, a 7-fold slower rate across rivers is responsible for an 11-month delay in the expected appearance of rabies in the southeast corner of Connecticut. The expected delay would be 16 months without long-distance translocation.
The early appearance of rabies in townships noncontiguous to infected townships could be caused by translocation events, delayed detection in adjacent townships, or even laboratory misdiagnosis. Although these causes could not be distinguished, we were able to discern whether the early occurrence of raccoon rabies in a township was temporally related to the development of a subsequent epidemic by regressing the time of the first case of raccoon rabies on the time to occurrence of the median observed case of rabies during the initial epidemic occurring in the township. Epidemics in the majority of townships developed rapidly after the first case of raccoon rabies was detected, reaching the median case after a delay of 4 months (Fig. 3c). By this measure, Putnam Township was an extreme outlier (Fig. 3c); the delay between the first observed case and the median case was out of proportion with the other cases. We repeated the fitting process, omitting Putnam Township from the analysis. Three townships, South Windsor, Union, and Enfield, were significantly early compared with the model, accounting for ≈16% of the total χ2 (Fig. 3 b and c).
A good statistical fit required the addition of habitat heterogeneity. The goodness of fit measure improved by ≈10% in the models that included rivers compared with those that did not (Table 1). This margin is much larger than the 1% improvement in the goodness of fit measure gained by incorporating human population density. The relatively large improvement suggests that rivers play an important role in rabies transmission. Moreover, when Putnam was excluded, models that did not incorporate rivers were rejected by the goodness of fit test, and those that did incorporate rivers were not. A comparison of the significance values, parameter values, and improvement in the goodness of fit from the addition of rivers suggests that a predictive model requires a 7-fold reduction in transmission crossing rivers (fitted parameters are reported in Table 1).
Although local transmission accounted for most transmission, long-distance translocation was frequent. Of the 159 townships not on the western border of Connecticut, 21 townships (13%) recorded their first case of raccoon rabies when none of the adjacent townships were infected.
Human population density was weakly associated with the rate of local spread and rate of translocation (the difference in the rate of spread between the least and most populated township was 11% in RivHum2 and 14% in RivHum1). The fits did improve when human population density was used to predict the rate of spread, but the improvements were minor, and the improved fit may be artifactual. Any association between the rate of spread and human population density is consistent with a hypothesis of increased rate of rabies spread into the townships, but it also supports a hypothesis of more efficient detection.
Most importantly, we found that river crossings slowed the spread of rabies by a factor of 7. The earliest appearance of rabies east of the Connecticut river may have been caused by a rare translocation event rather than a more predictable local transmission event; the first case of rabies east of the Connecticut river was detected in South Windsor (sw in Figs. 1 and 3b), far in advance of the front, and the first cases in the nearby townships of Enfield and Union and one adjacent township were observed shortly thereafter (e and u in Figs. 1 and 3b). Similar but less dramatic patterns are observed in the southern, central townships surrounding Clinton (c in Figs. 1 and 3b).
To assess the consequences of a 7-fold delay crossing rivers on the overall dynamics of rabies, we simulated the epidemic further with and without rivers and with and without long-distance translocation. Rivers delay the appearance in southeastern Connecticut by ≈16 months (Fig. 3d) without translocation and by 11 months with translocation. The effect of rivers was strongest close to the river and declined with the distance from the river. A river shadow effect was visible (Fig. 3d). We attribute this shadow effect to two properties of these models. First, a small probability of translocation adds stochasticity that obscures the patterns that would be formed by local heterogeneity. Local delays associated with river crossings are reduced if rabid raccoons cross rivers by translocation. Second, patterns caused by local heterogeneity dissipate, because there are multiple routes for the spread of infection. For example, once an epidemic reaches a river, it may cross at any of the shared boundaries. After the epidemic crosses the river, it may spread rapidly down the river to other townships. Consequently, a long local delay crossing the river may appear shorter, because it is affected most strongly by the average first event, not the average of all events.
Discussion
The local slowing in the rate of spatial propagation caused by rivers can be compared with the intended effect of a cordon sanitaire such as might be achieved by distributing rabies vaccine-laden baits intended for consumption by raccoons (17–19). The strategy of distributing oral vaccines for raccoons is currently being employed in Florida, Maryland, Massachusetts, New Hampshire, New York, Ohio, and Virginia and is planned for use in several other states (20, 21). Diffusion models for the spatial spread of rabies among foxes suggested that control zones might prevent the spread of rabies into new areas, and estimated the width of a cordon sanitaire necessary to stop the spread (3). Subsequent models developed the mathematical theory of invasive spread, emphasizing the role of chance long-distance translocation that can change the rate of spread (1, 4, 22, 23). Our analysis suggests that translocation of rabid raccoons represents a serious threat to management strategies designed to contain the spatial spread of rabies (24–26). The repeated translocation of rabid animals into areas in which the disease is absent is likely to establish new foci even if every introduction does not seed an epidemic. To be most effective, an attempt to control the spread of rabies should be accompanied by active surveillance to detect new foci in regions beyond the borders of the cordon sanitaire.
Predicting infectious disease emergence depends on being able to answer three basic questions. First, where will the disease emerge? Second, once established is there a predictable time course for the frequency and duration of reemergent epidemics? Third, from the initial point of emergence, where and how will the disease spread? The first of these questions seems difficult. As in the case for the mid-Atlantic rabies epidemic among raccoons, the point of initiation was determined by an unpredictable event, the translocation of rabid animals into a previously unexposed population. Many cases of recent disease emergence can be associated with similar unpredictable events, as exemplified by the emergence of West Nile virus in the northeast U.S. (27) and foot and mouth disease in the United Kingdom (28). Analytic models for predicting the temporal and spatial occurrence of infectious disease once established, however, may be more amenable to construction (29).
Elsewhere we have described an algorithmic approach to reconstructing and predicting the temporal pattern of rabies epidemics in raccoons (30). Here we present a different methodology for linking large databases, geographic information systems, and stochastic simulations in constructing predictive probabilistic models of spatial dynamics. Similar approaches have been used to evaluate how multiple factors influence the spatial dynamics and incidence of measles in Bristol, U.K. (31). We suggest that simple models are powerful tools to detect heterogeneous spread in observational data. Our analysis suggests that incorporating environmental spatial heterogeneity and risk estimates for long-distance translocation are critical for predicting the spread of infection across geographic regions.
The model we have proposed, although predicting a substantial portion of the data, leaves unaccounted the long delay to appearance of raccoon rabies in clustered subsets of the townships in southeastern and northwestern Connecticut (Fig. 3b). We suggest that these delays may be caused by additional environmental spatial variables that were not included in this analysis. In particular, the disease seems to have traveled northeast, avoiding northwestern Connecticut and arcing past the Connecticut River prior to spreading into the southeast. We hypothesize that these patterns of movement may be associated with other measures of raccoon habitat and physiographic barriers that form corridors for movement such as coniferous forests and mountains. We suspect that these patterns may emerge from simulations on heterogeneous landscapes using ecologically defined patches with a finer spatial resolution of the landscape. Environmental variables such as human population density may provide more information when the spatial distribution of humans is modeled at a finer spatial resolution than the township average.
The methods we describe here, because they pin-point specific spatial locations at which there is disagreement between the model and the data, can be used as a means of targeting new hypotheses on the role of specific potential environmental factors in influencing spatial patterns of disease emergence across geographic ranges. These methods should prove valuable in modeling disease dynamics where local propagation is affected by local geography, and we hope that others may find this approach useful for application to other infectious disease systems.
Acknowledgments
We thank Colin Russel, Charles Rupprecht, Aaron Curns, and John O'Connor for their comments on the paper. We also thank Matt Carter and James Hadler of the Connecticut Department of Health and other state epidemiologists and state laboratory personnel in the U.S. who have acquired and contributed these data over many years. This research has been supported by National Institutes of Health Grant R01 AI47498-01 (to L.A.R. and J.E.C.) and through the National Center for Ecological Analysis and Synthesis (a center funded by National Science Foundation Grant DEB-94-21535, the University of California, Santa Barbara, the California Resources Agency, and the California Environmental Protection Agency).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 3365.
References
- 1.Mollison D. Philos Trans R Soc London B. 1986;314:675–693. [Google Scholar]
- 2.Plowright W. Rev Sci Tech. 1988;7:783–795. doi: 10.20506/rst.7.4.380. [DOI] [PubMed] [Google Scholar]
- 3.Murray J D, Stanley E A, Brown D L. Proc R Soc London B. 1986;229:111–150. doi: 10.1098/rspb.1986.0078. [DOI] [PubMed] [Google Scholar]
- 4.Mollison D. Math Biosci. 1991;107:255–287. doi: 10.1016/0025-5564(91)90009-8. [DOI] [PubMed] [Google Scholar]
- 5.Shigaseda N. In: Population Biology, Morphogenesis and Neurosciences: Lecture Notes in Biomathematics. Teramoto E, Yamaguti M, editors. Vol. 71. Heidelberg: Springer; 1987. pp. 88–97. [Google Scholar]
- 6.Wilson M L, Bretsky P M, Cooper G H, Egbertson S H, Kruiningen H J V, Carter M L. Am J Trop Med Hyg. 1997;57:457–463. doi: 10.4269/ajtmh.1997.57.457. [DOI] [PubMed] [Google Scholar]
- 7.Krebs J W, Holman R C, Hines U, Strine T, Mandel E, Childs J E. J Am Vet Med Assoc. 1992;201:1836–1848. [PubMed] [Google Scholar]
- 8.Nettles V F, Shaddock J H, Sikes R K, Reyes C R. Am J Public Health. 1979;69:601–602. doi: 10.2105/ajph.69.6.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins S R, Winkler W G. Am J Epidemiol. 1987;126:429–437. doi: 10.1093/oxfordjournals.aje.a114674. [DOI] [PubMed] [Google Scholar]
- 10.Smith J S, Sumner J W, Roumillat L F, Baer G M, Winkler W G. J Infect Dis. 1984;149:769–774. doi: 10.1093/infdis/149.5.769. [DOI] [PubMed] [Google Scholar]
- 11.Smith J S, Yager P A, Bigler W J, Hartwig E C J. J Wildl Dis. 1990;26:473–485. doi: 10.7589/0090-3558-26.4.473. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman C O, Gottschang J L. J Mammal. 1997;56:623–636. [Google Scholar]
- 13.Riley S P D, Hadidian J, Manski D A. Can J Zool. 1998;76:1153–1164. [Google Scholar]
- 14.Carey A B, Giles R H, McLean R G. Am J Trop Med Hyg. 1978;27:573–580. doi: 10.4269/ajtmh.1978.27.573. [DOI] [PubMed] [Google Scholar]
- 15.Moore D A. Prev Vet Med. 1999;40:19–32. doi: 10.1016/s0167-5877(99)00005-7. [DOI] [PubMed] [Google Scholar]
- 16.Lotze J H, Anderson S. Mamm Species. 1979;119:1–8. [Google Scholar]
- 17.Hadidian J, Jenkins S R, Johnston D H. J Wildl Dis. 1989;25:1–9. doi: 10.7589/0090-3558-25.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Hanlon C A, Niezgoda M, Hamir A N. J Wildl Dis. 1998;34:170–179. doi: 10.7589/0090-3558-34.2.228. [DOI] [PubMed] [Google Scholar]
- 19.Rupprecht C E, Wiktor T J, Johnson D J, Hamir A N, Tietzschold B, Wunner W H, Glickman L T, Koprowski H. Proc Natl Acad Sci USA. 1986;83:7947–7950. doi: 10.1073/pnas.83.20.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanlon C A, Rupprecht C E. In: Emerging Infections. Scheld W M, Armstrong D, Hughes J M, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 59–80. [Google Scholar]
- 21.Robbins A H, Borden M D, Windmiller B S, Niezgoda M, Marcus L C, O'Brien S M, Kreindel S M, McGuill M W, DeMaria A J, Rupprecht C E, Rowell S. J Am Vet Med Assoc. 1998;213:1407–1412. [PubMed] [Google Scholar]
- 22.Shigaseda N, Kawasaki K, Takeda Y. Am Nat. 1995;146:229–251. [Google Scholar]
- 23.Lewis M, Pacala S W. Am Nat. 1995;146:229–251. [Google Scholar]
- 24.Kallen A, Arcuri P, Murray J D. J Theor Biol. 1985;116:377–393. doi: 10.1016/s0022-5193(85)80276-9. [DOI] [PubMed] [Google Scholar]
- 25.Smith G C, Harris S. Philos Trans R Soc London B. 1991;334:459–479. doi: 10.1098/rstb.1991.0127. [DOI] [PubMed] [Google Scholar]
- 26.Evans N D, Pritchard A J. IMA J Math Appl Med Biol. 2001;18:1–23. [PubMed] [Google Scholar]
- 27.Garmendia A E, Van Kruiningen H J, French R A. Microbes Infect. 2001;3:223–229. doi: 10.1016/s1286-4579(01)01374-0. [DOI] [PubMed] [Google Scholar]
- 28.Pickrell J, Enserink M. Science. 2001;291:1677. doi: 10.1126/science.291.5509.1677a. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson N M, Donnelly C A, Anderson R M. Science. 2001;292:1155–1160. doi: 10.1126/science.1061020. [DOI] [PubMed] [Google Scholar]
- 30.Childs J E, Curns A T, Dey M E, Real L A, Feinstein L, Bjornstad O N, Krebs J W. Proc Natl Acad Sci USA. 2000;97:13666–13671. doi: 10.1073/pnas.240326697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haggett P. Econ Geogr. 1976;52:136–146. [Google Scholar]