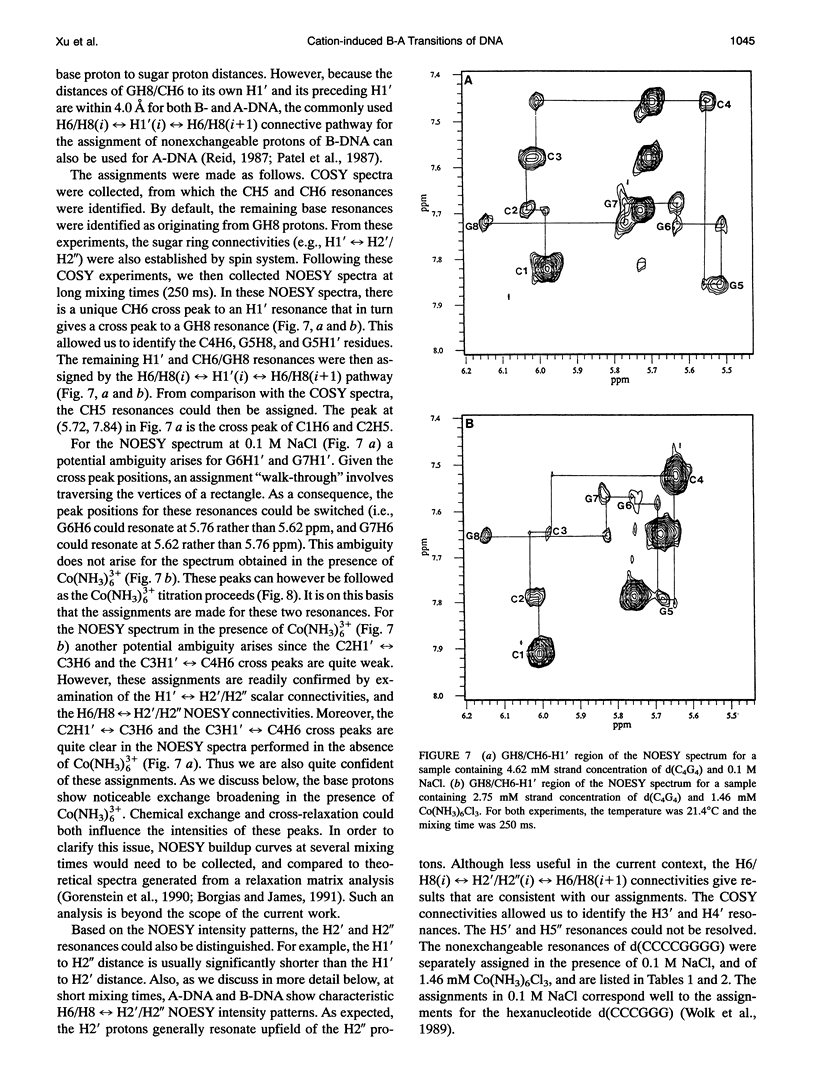
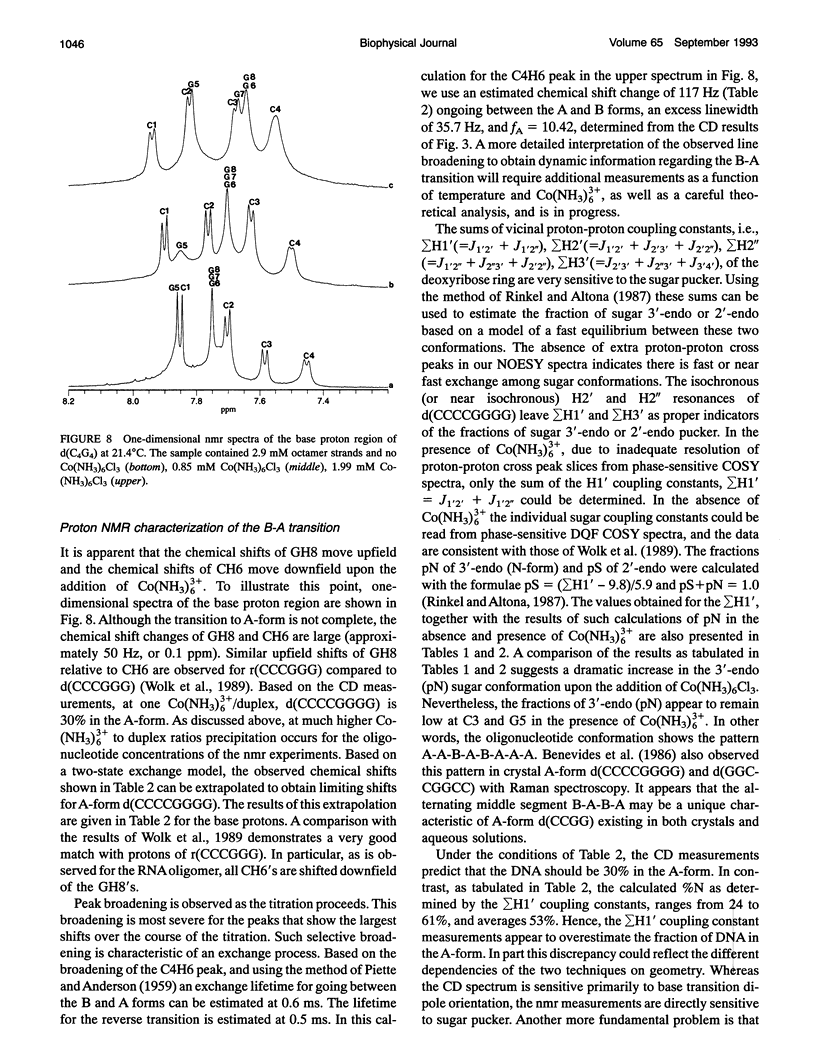
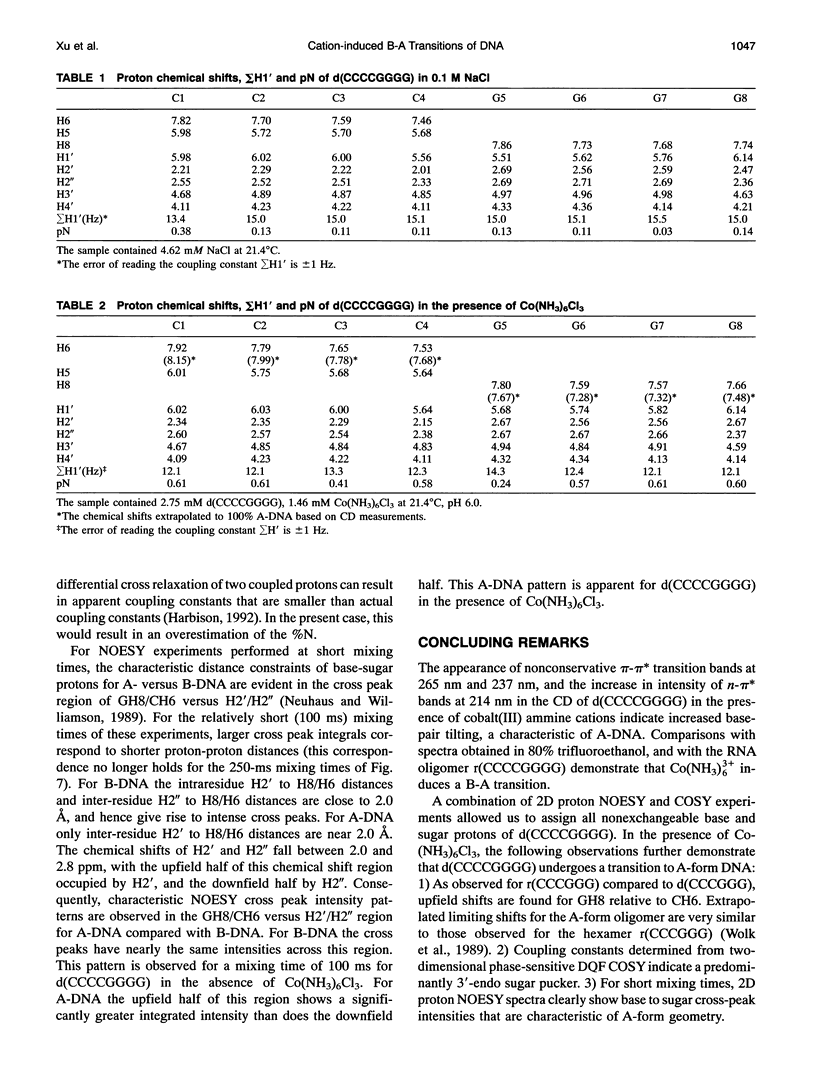
Abstract
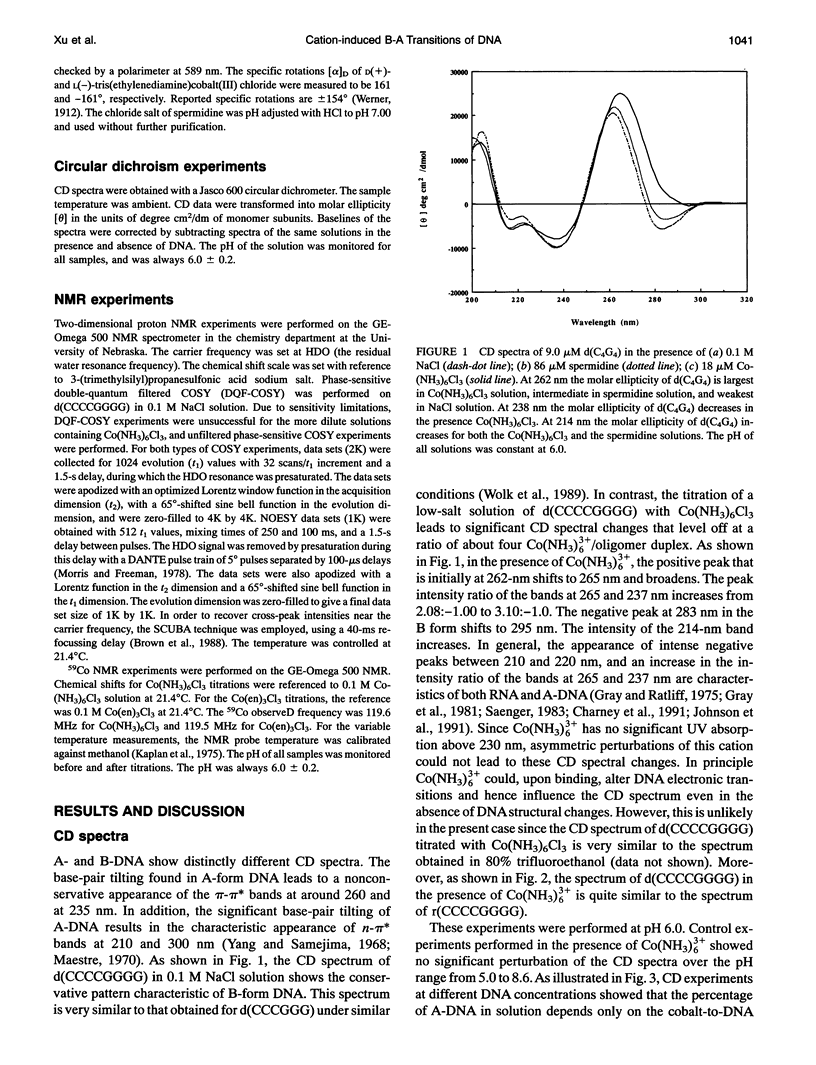
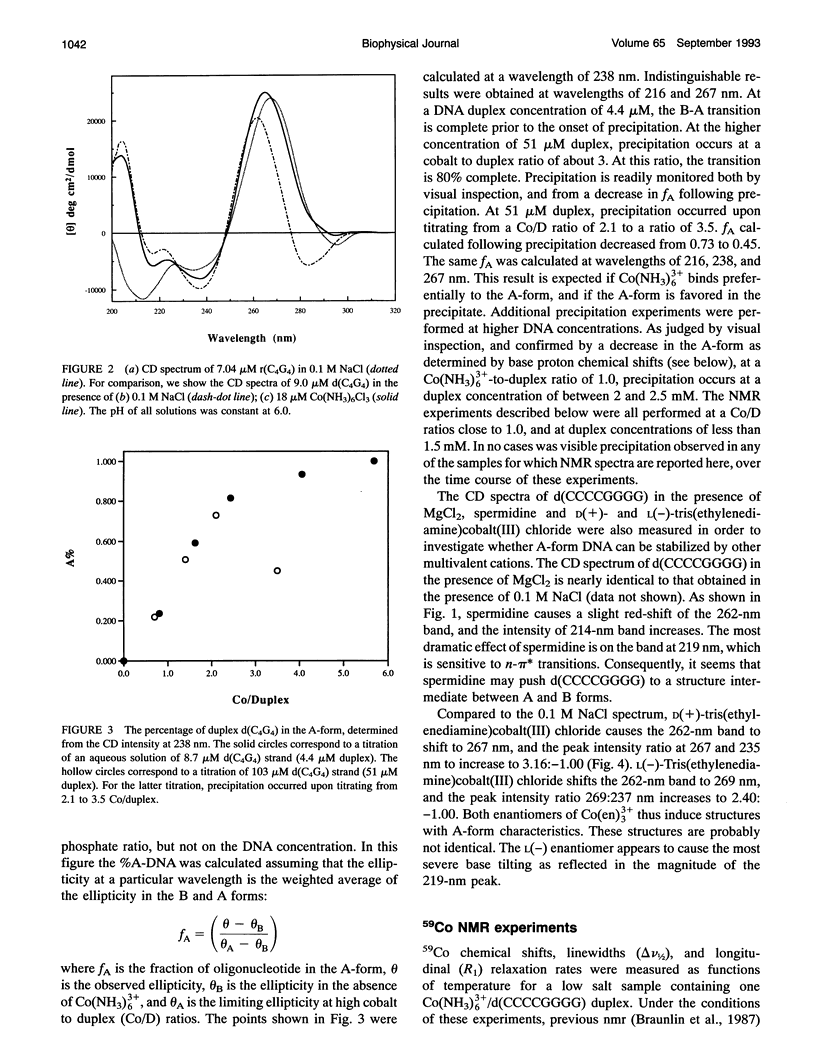
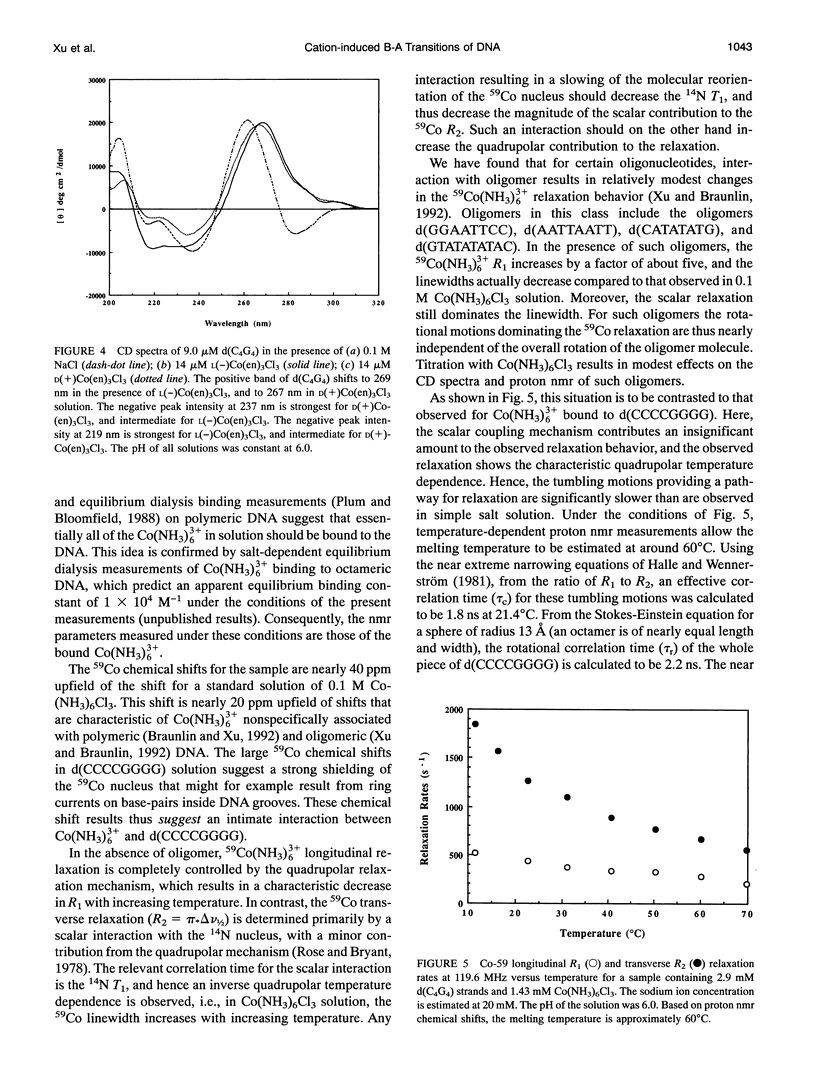
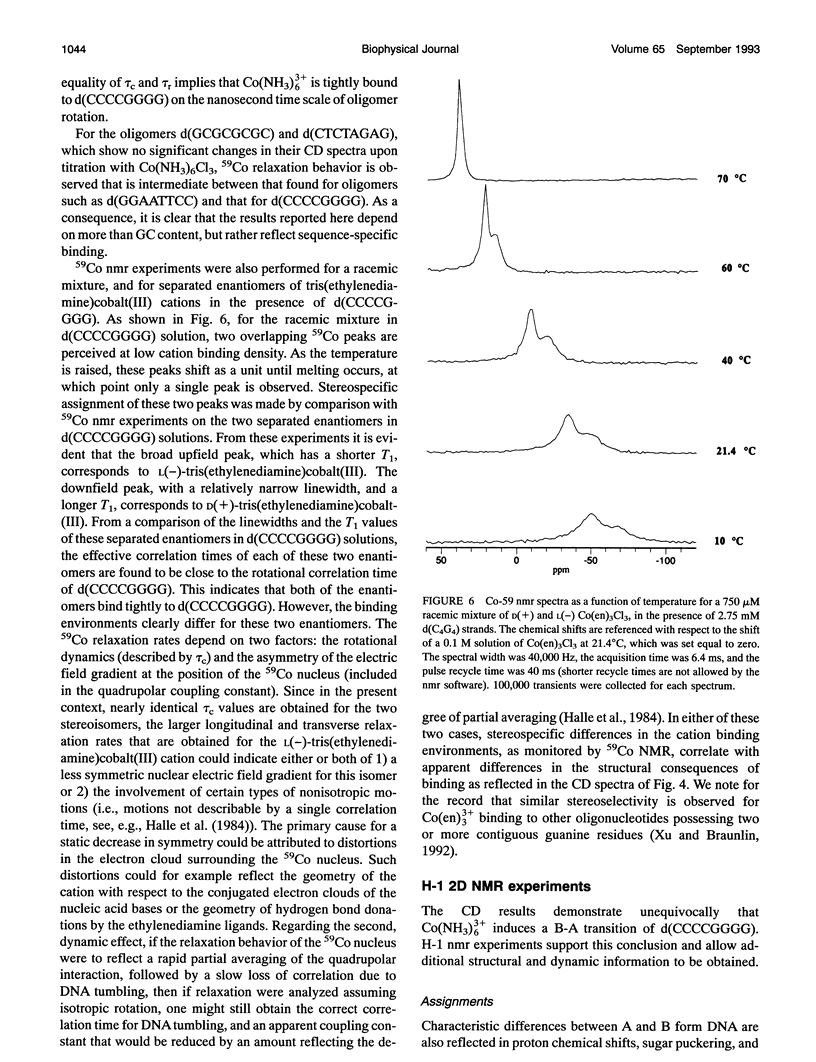
Circular dichroism (CD) spectra of d(CCCCGGGG) in the presence of Co(NH3)6(3+) are very similar to spectra of r(CCCCGGGG). In contrast, B-form characteristics are observed for d(CCCCGGGG) in the presence of Na+ and Mg2+, even at high salt concentrations. Spermidine induces modest changes of the CD of d(CCCCGGGG). The NMR chemical shifts of the nonexchangeable protons of d(CCCCGGGG) in the absence and presence of Co(NH3)6(3+) were assigned by proton two-dimensional (2D) NOESY and COSY measurements. The chemical shifts of the GH8 protons of d(CCCCGGGG) move upfield upon titration with Co(NH3)6Cl3. The sums of the sugar H1' coupling constants decrease with added Co(NH3)6Cl3. Cross peak intensities in the 2D proton NOESY spectra show a transformation from B-DNA to A-DNA characteristics upon the addition of Co(NH3)6Cl3. The temperature-dependent 59Co transverse and longitudinal relaxation rates demonstrate that Co(NH3)6(3+) is site-bound to the oligomer. Such localization is not a general feature of Co(NH3)6(3+) binding to oligonucleotides. 59Co NMR relaxation and CD measurements demonstrate chiral discrimination by d(CCCCGGGG) for the two stereoisomers of Co(en)3(3+). Both stereoisomers bind tightly as judged by 59Co NMR, and both cause large (but nonequivalent) changes in the CD of this oligomer.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Varani G., Walker G. T., Tinoco I., Jr The TFIIIA recognition fragment d(GGATGGGAG).d(CTCCCATCC) is B-form in solution. Nucleic Acids Res. 1988 Apr 25;16(8):3559–3572. doi: 10.1093/nar/16.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton J. K. Metals and DNA: molecular left-handed complements. Science. 1986 Aug 15;233(4765):727–734. doi: 10.1126/science.3016894. [DOI] [PubMed] [Google Scholar]
- Benevides J. M., Wang A. H., Rich A., Kyogoku Y., van der Marel G. A., van Boom J. H., Thomas G. J., Jr Raman spectra of single crystals of r(GCG)d(CGC) and d(CCCCGGGG) as models for A DNA, their structure transitions in aqueous solution, and comparison with double-helical poly(dG).poly(dC). Biochemistry. 1986 Jan 14;25(1):41–50. doi: 10.1021/bi00349a007. [DOI] [PubMed] [Google Scholar]
- Braunlin W. H., Anderson C. F., Record M. T., Jr Competitive interactions of Co(NH3)6(3+) and Na+ with helical B-DNA probed by 59Co and 23Na NMR. Biochemistry. 1987 Dec 1;26(24):7724–7731. doi: 10.1021/bi00398a028. [DOI] [PubMed] [Google Scholar]
- Braunlin W. H., Xu Q. Hexaamminecobalt(III) binding environments on double-helical DNA. Biopolymers. 1992 Dec;32(12):1703–1711. doi: 10.1002/bip.360321212. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Warshaw M. M., Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9(9):1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- Charney E., Chen H. H., Rau D. C. The flexibility of A-form DNA. J Biomol Struct Dyn. 1991 Oct;9(2):353–362. doi: 10.1080/07391102.1991.10507917. [DOI] [PubMed] [Google Scholar]
- Fairall L., Martin S., Rhodes D. The DNA binding site of the Xenopus transcription factor IIIA has a non-B-form structure. EMBO J. 1989 Jun;8(6):1809–1817. doi: 10.1002/j.1460-2075.1989.tb03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M., Blanco J., Tennant L. L. The 5S gene internal control region is B-form both free in solution and in a complex with TFIIIA. Nature. 1987 Oct 1;329(6138):460–462. doi: 10.1038/329460a0. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Ratliff R. L. Circular dichroism spectra of poly[d(AC):d(GT)], poly[r(AC):r(GU)], and hybrids poly[d(AC):r(GU)] and poly[r(AC):d(GT)] in the presence of ethanol. Biopolymers. 1975 Mar;14(3):487–498. doi: 10.1002/bip.1975.360140305. [DOI] [PubMed] [Google Scholar]
- Haran T. E., Shakked Z., Wang A. H., Rich A. The crystal structure of d(CCCCGGGG): a new A-form variant with an extended backbone conformation. J Biomol Struct Dyn. 1987 Oct;5(2):199–217. doi: 10.1080/07391102.1987.10506390. [DOI] [PubMed] [Google Scholar]
- Hingerty B. E., Brown R. S., Klug A. Stabilization of the tertiary structure of yeast phenylalanine tRNA by [Co(NH3)6]3+. X-ray evidence for hydrogen bonding to pairs of guanine bases in the major groove. Biochim Biophys Acta. 1982 Apr 26;697(1):78–82. doi: 10.1016/0167-4781(82)90047-1. [DOI] [PubMed] [Google Scholar]
- Huber P. W., Morii T., Mei H. Y., Barton J. K. Structural polymorphism in the major groove of a 5S RNA gene complements the zinc finger domains of transcription factor IIIA. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10801–10805. doi: 10.1073/pnas.88.23.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. H., Gray D. M., Sutherland J. C. Vacuum UV CD spectra of homopolymer duplexes and triplexes containing A.T or A.U base pairs. Nucleic Acids Res. 1991 May 11;19(9):2275–2280. doi: 10.1093/nar/19.9.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Maestre M. F. Circular dichroism of DNA films: reversibility studies. J Mol Biol. 1970 Sep 28;52(3):543–556. doi: 10.1016/0022-2836(70)90418-3. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Torigoe C., Tsuboi M. An A-form poly(dG).poly(dC) in H2O solution. Biopolymers. 1985 Sep;24(9):1841–1844. doi: 10.1002/bip.360240913. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Hare D. DNA and RNA: NMR studies of conformations and dynamics in solution. Q Rev Biophys. 1987 Aug;20(1-2):35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Plum G. E., Bloomfield V. A. Equilibrium dialysis study of binding of hexammine cobalt(III) to DNA. Biopolymers. 1988 Jun;27(6):1045–1051. doi: 10.1002/bip.360270611. [DOI] [PubMed] [Google Scholar]
- Reid B. R. Sequence-specific assignments and their use in NMR studies of DNA structure. Q Rev Biophys. 1987 Aug;20(1-2):1–34. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Rinkel L. J., Altona C. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. J Biomol Struct Dyn. 1987 Feb;4(4):621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Appella E., Omichinski J. G., Clore G. M., Gronenborn A. M. Specific DNA binding to a major histocompatibility complex enhancer sequence by a synthetic 57-residue double zinc finger peptide from a human enhancer binding protein. J Biol Chem. 1991 Apr 15;266(11):7306–7311. [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Wolk S., Thurmes W. N., Ross W. S., Hardin C. C., Tinoco I., Jr Conformational analysis of d(C3G3), a B-family duplex in solution. Biochemistry. 1989 Mar 21;28(6):2452–2459. doi: 10.1021/bi00432a016. [DOI] [PubMed] [Google Scholar]
- Yang J. T., Samejima T. Effect of base tilting on the optical activity of nucleic acids: a hypothesis. Biochem Biophys Res Commun. 1968 Dec 9;33(5):739–745. doi: 10.1016/0006-291x(68)90221-0. [DOI] [PubMed] [Google Scholar]
- You Q. M., Veldhoen N., Baudin F., Romaniuk P. J. Mutations in 5S DNA and 5S RNA have different effects on the binding of Xenopus transcription factor IIIA. Biochemistry. 1991 Mar 5;30(9):2495–2500. doi: 10.1021/bi00223a028. [DOI] [PubMed] [Google Scholar]


