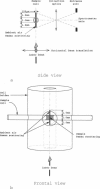
Abstract
The bond stretching vibration of the cysteine sulfhydryl (SH) group in a typical protein generates a Raman band in the spectral interval 2500-2600 cm-1, a region devoid of interference from any other fundamental mode of vibration of the protein. The relatively high Raman cross section associated with the S-H stretching vibration, the sensitivity of the vibrational frequency to hydrogen bonding interactions and side chain configurations, and the dependence of the Raman intensity on thiol-thiolate equilibria, combine to make the Raman SH band a potentially valuable marker of protein sulfhydryl interactions and a unique indicator of sulfhydryl participation in thiol-disulfide oxidoreductase activity. In order to exploit Raman spectroscopy for these purposes, accurate and precise measurements of Raman SH band profiles are required. We show here that the required precision and accuracy can be achieved by use of the Raman band corresponding to the stretching vibration of in situ nitrogen gas as a quantitative intensity and frequency standard. The Raman Q-branch center of the N2 band occurs at 2330.7 cm-1.
Full text
PDF
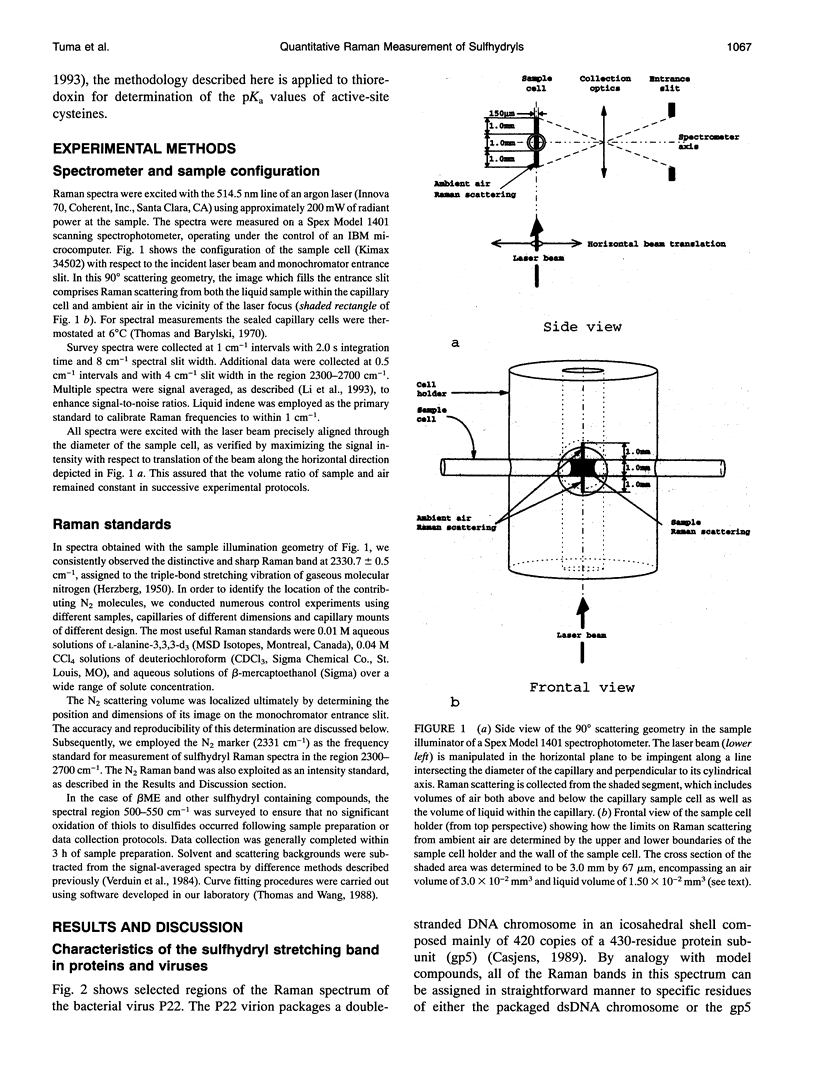
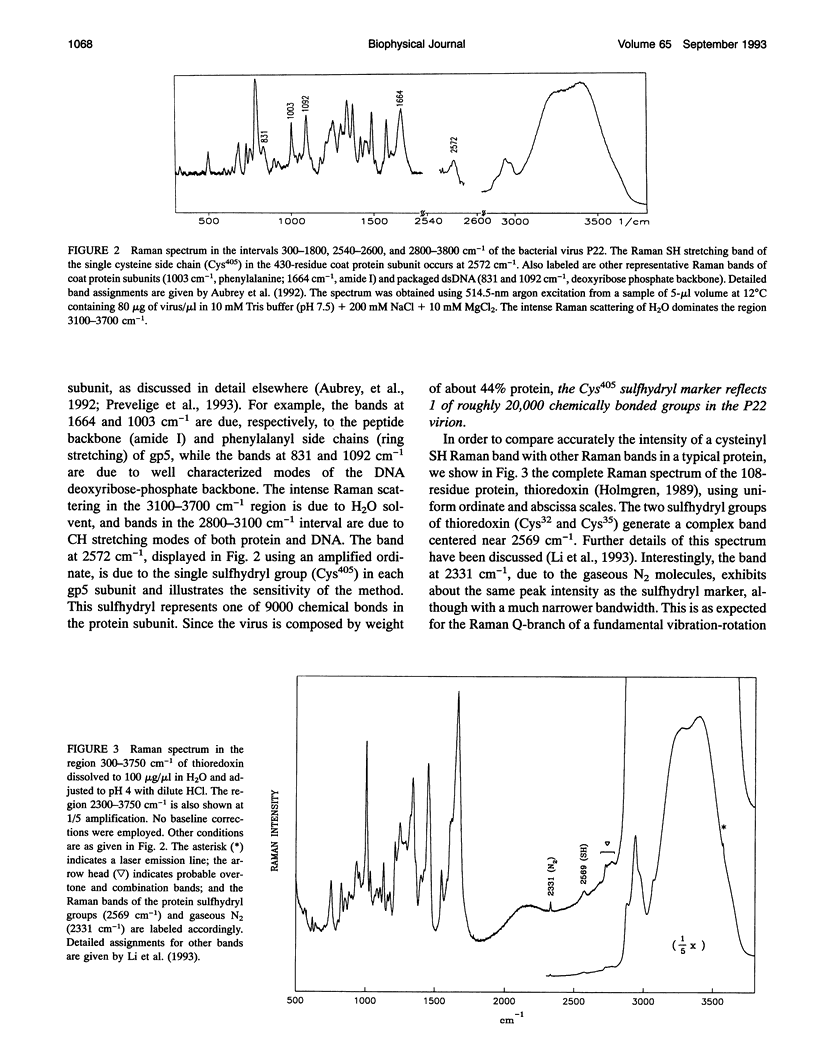
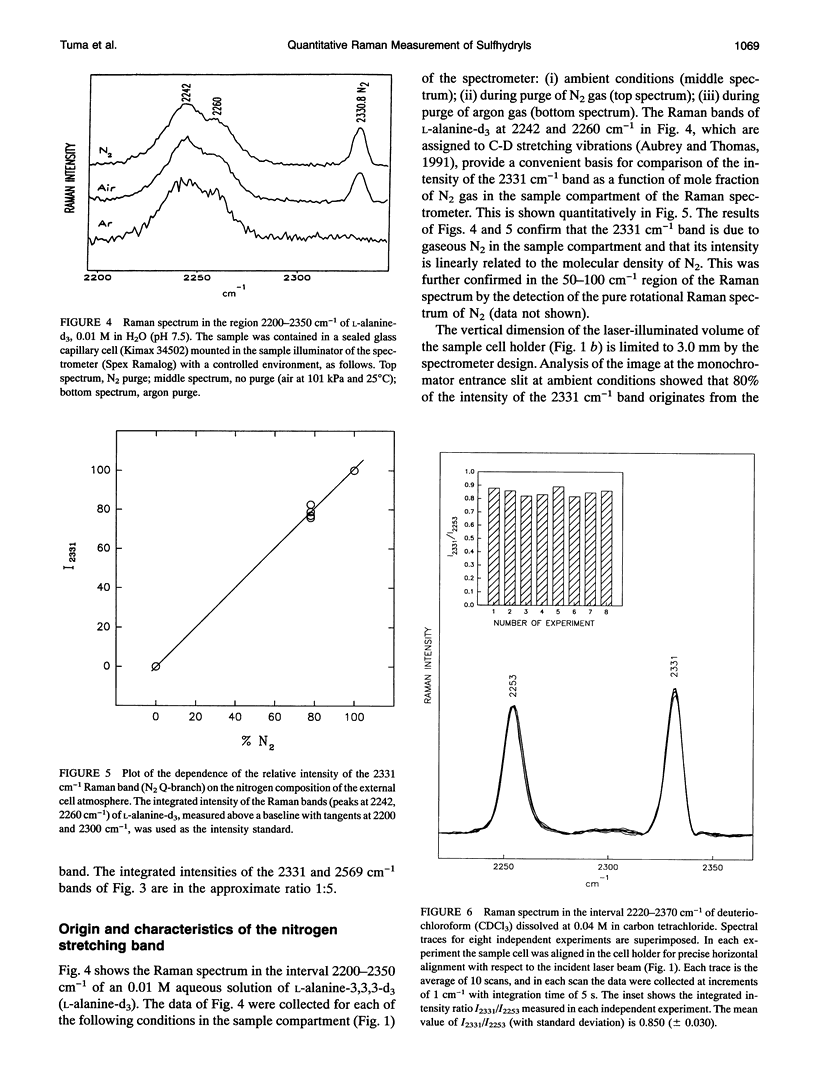

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
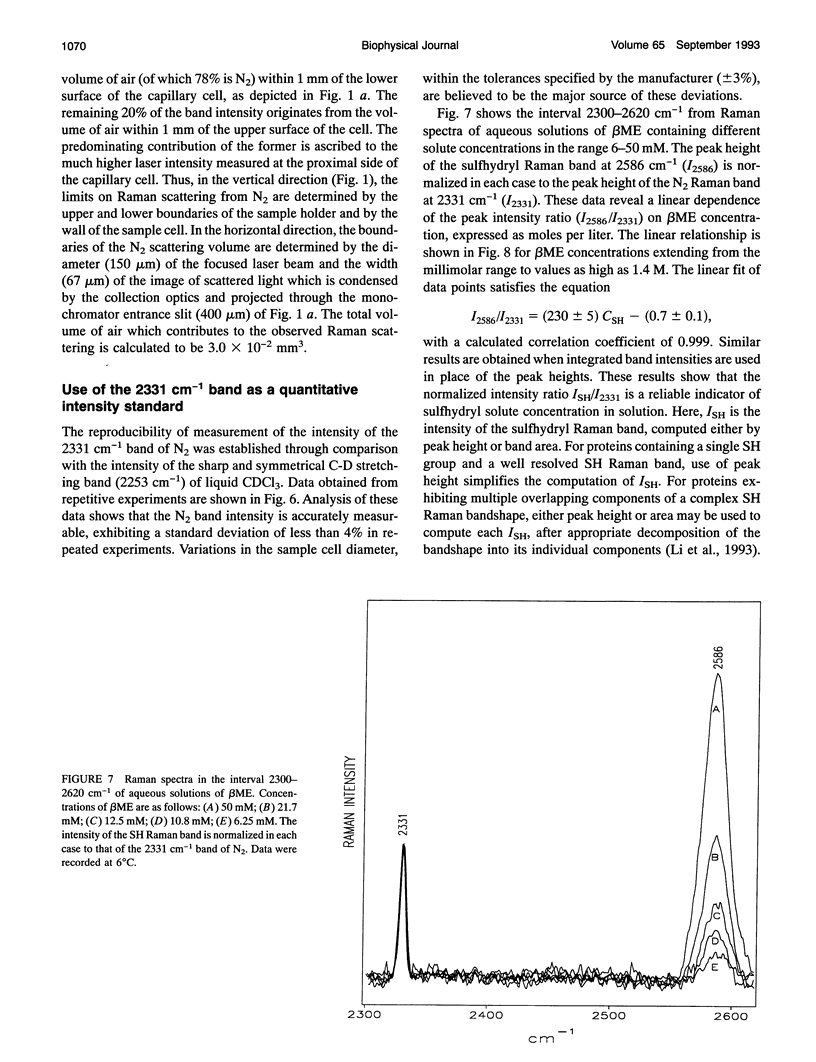
- Aubrey K. L., Casjens S. R., Thomas G. J., Jr Secondary structure and interactions of the packaged dsDNA genome of bacteriophage P22 investigated by Raman difference spectroscopy. Biochemistry. 1992 Dec 1;31(47):11835–11842. doi: 10.1021/bi00162a023. [DOI] [PubMed] [Google Scholar]
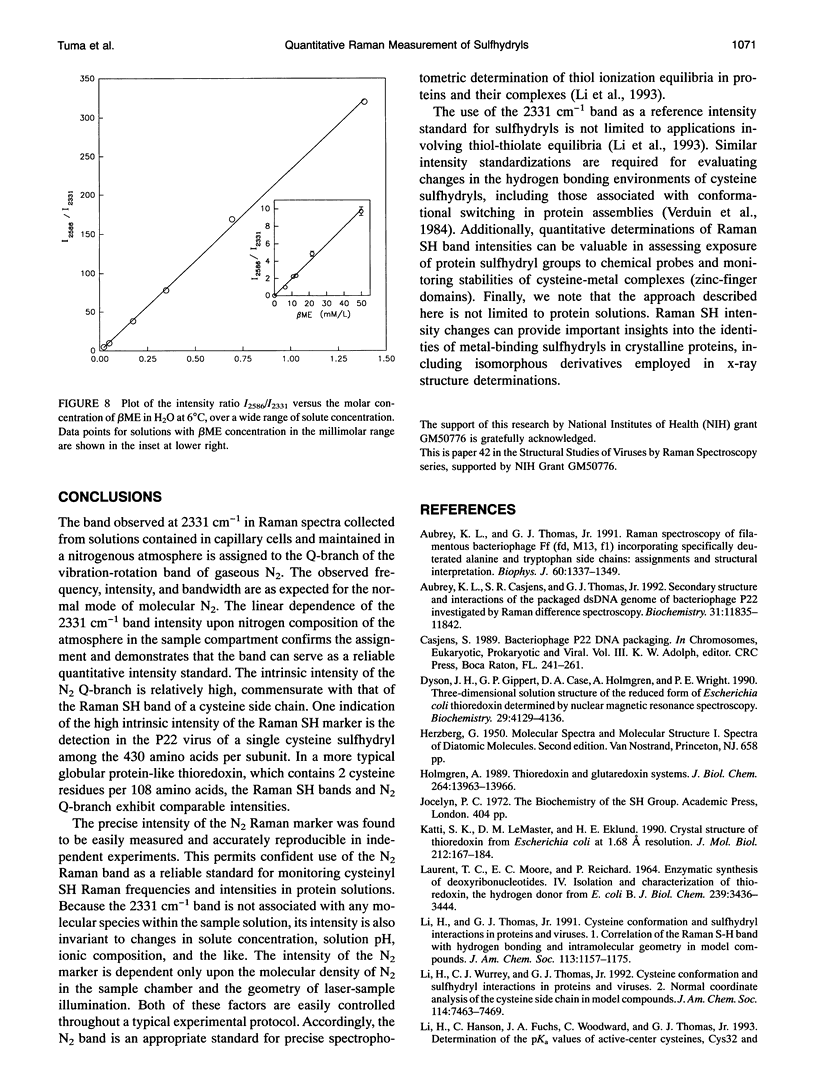
- Aubrey K. L., Thomas G. J., Jr Raman spectroscopy of filamentous bacteriophage Ff (fd, M13, f1) incorporating specifically-deuterated alanine and tryptophan side chains. Assignments and structural interpretation. Biophys J. 1991 Dec;60(6):1337–1349. doi: 10.1016/S0006-3495(91)82171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J., Gippert G. P., Case D. A., Holmgren A., Wright P. E. Three-dimensional solution structure of the reduced form of Escherichia coli thioredoxin determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1990 May 1;29(17):4129–4136. doi: 10.1021/bi00469a016. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989 Aug 25;264(24):13963–13966. [PubMed] [Google Scholar]
- Katti S. K., LeMaster D. M., Eklund H. Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution. J Mol Biol. 1990 Mar 5;212(1):167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- LAURENT T. C., MOORE E. C., REICHARD P. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEOTIDES. IV. ISOLATION AND CHARACTERIZATION OF THIOREDOXIN, THE HYDROGEN DONOR FROM ESCHERICHIA COLI B. J Biol Chem. 1964 Oct;239:3436–3444. [PubMed] [Google Scholar]
- Prevelige P. E., Jr, Thomas D., Aubrey K. L., Towse S. A., Thomas G. J., Jr Subunit conformational changes accompanying bacteriophage P22 capsid maturation. Biochemistry. 1993 Jan 19;32(2):537–543. doi: 10.1021/bi00053a019. [DOI] [PubMed] [Google Scholar]
- Verduin B. J., Prescott B., Thomas G. J., Jr RNA-protein interactions and secondary structures of cowpea chlorotic mottle virus for in vitro assembly. Biochemistry. 1984 Sep 11;23(19):4301–4308. doi: 10.1021/bi00314a008. [DOI] [PubMed] [Google Scholar]