Abstract
Skeletal muscle myosin is an enzyme that interacts allosterically with MgATP and actin to transduce the chemical energy from ATP hydrolysis into work. By modifying myosin structure, one can change this allosteric interaction and gain insight into its mechanism. Chemical cross-linking with N,N'-p-phenylenedimaleimide (pPDM) of Cys-697 to Cys-707 of the myosin-ADP complex eliminates activity and produces a species that resembles myosin with ATP bound (Burke et al., 1976). Nucleotide-free pPDM-modified myosin subfragment 1 (S1) was prepared, and its structural and allosteric properties were investigated by comparing the nucleotide and actin interactions of S1 to those of pPDM-S1. The structural properties of the nucleotide-free pPDM-S1 are different from those of S1 in several respects. pPDM-S1 intrinsic tryptophan fluorescence intensity is reduced 28%, indicating a large increase of an internal quenching reaction (the fluorescence intensity of the related vanadate complex of S1, S1-MgADP-Vi, is reduced by a similar degree). Tryptophan fluorescence anisotropy increases from 0.168 for S1 to 0.192 for pPDM-S1, indicating that the unquenched tryptophan population in pPDM-S1 has reduced local freedom of motion. The actin affinity of pPDM-S1 is over 6,000-fold lower than that of S1, and the absolute value of the product of the net effective electric charges at the acto-S1 interface is reduced from 8.1 esu2 for S1 to 1.6 esu2 for pPDM-S1. In spite of these changes, the structural response of pPDM-S1 to nucleotide and the allosteric communication between its ATP and actin sites remain intact. Compared to pPDM-S1, the fluorescence intensity of pPDM-S1 *MgADP is increased 50%(compared to 8 and 31% increases, respectively, for MgADP and MgATP binding to S1). Compared to acto-pPDM-S1, the absolute value of the product of the net effective electric charge at the actin binding interface of acto-pPDM-S1 *MgADP increases 7.3 esu2 (compared to a 0.9 esu2 decrease and an 11.0 esu2 increase, respectively, for MgADP and MgATP binding to acto-Sl).The interaction free energy for the ligands MgADP and actin, is -2.0 kcal/mol for pPDM-S1, compared to -1.2 kcal/mol for unmodified S1.
Full text
PDF

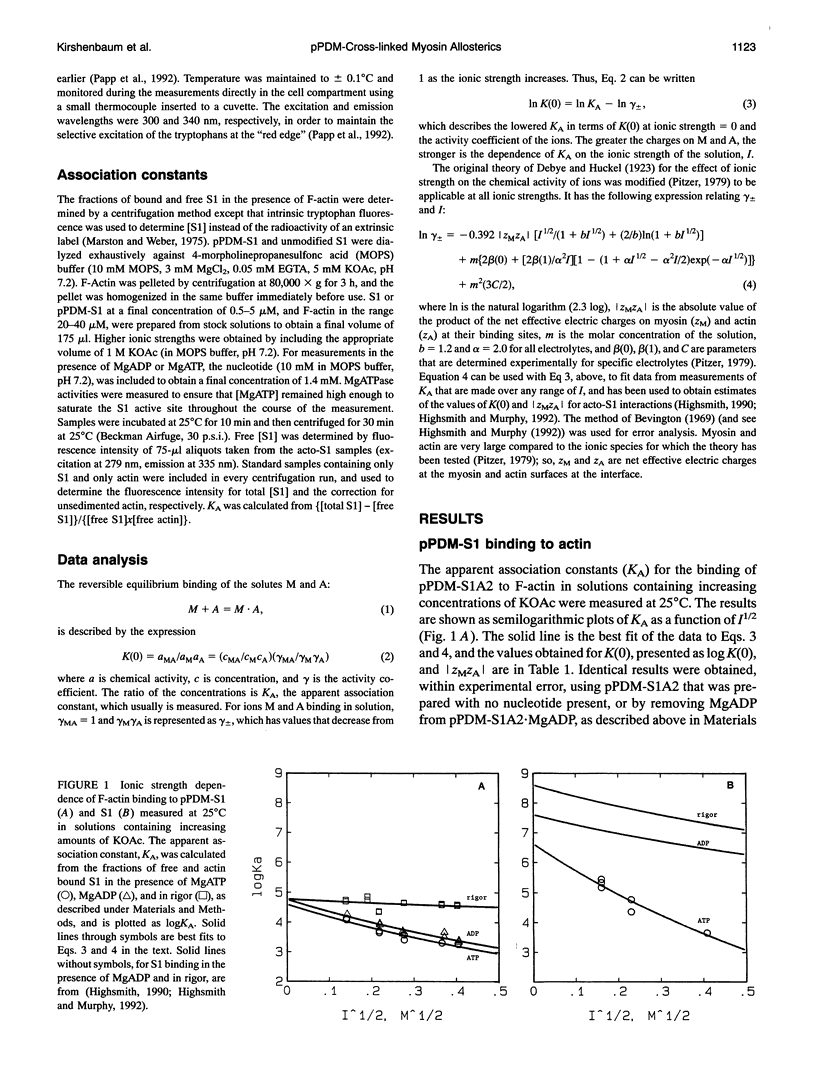
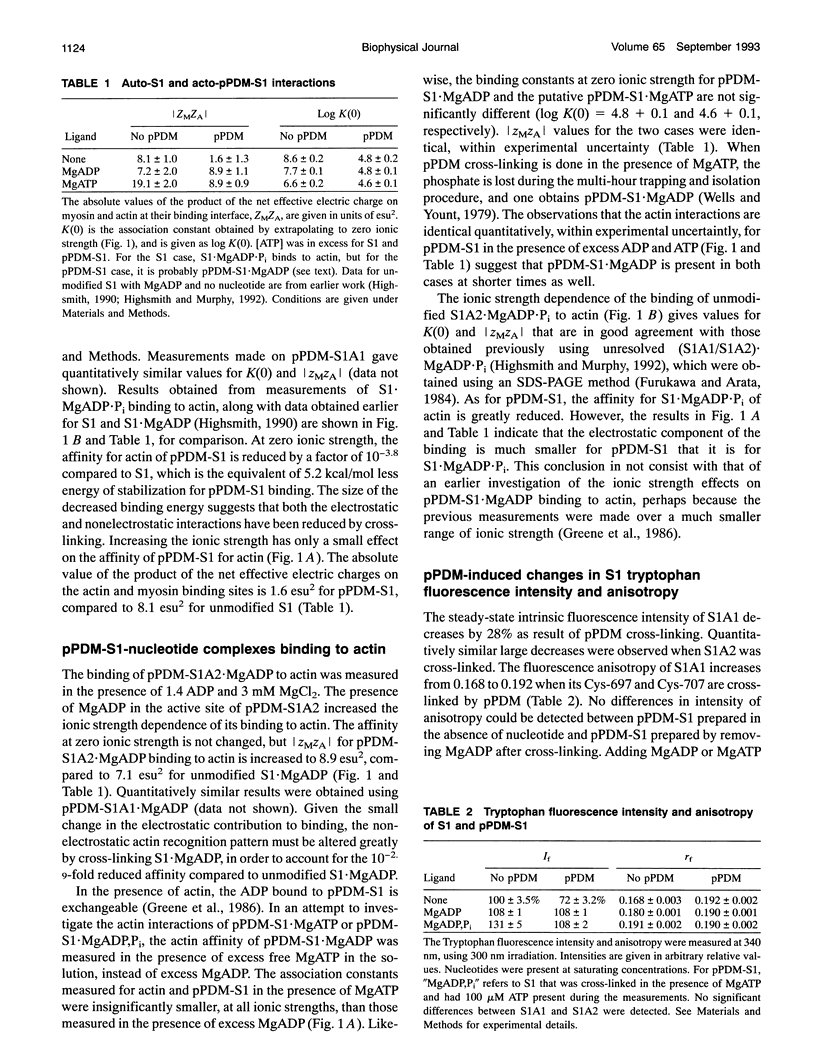
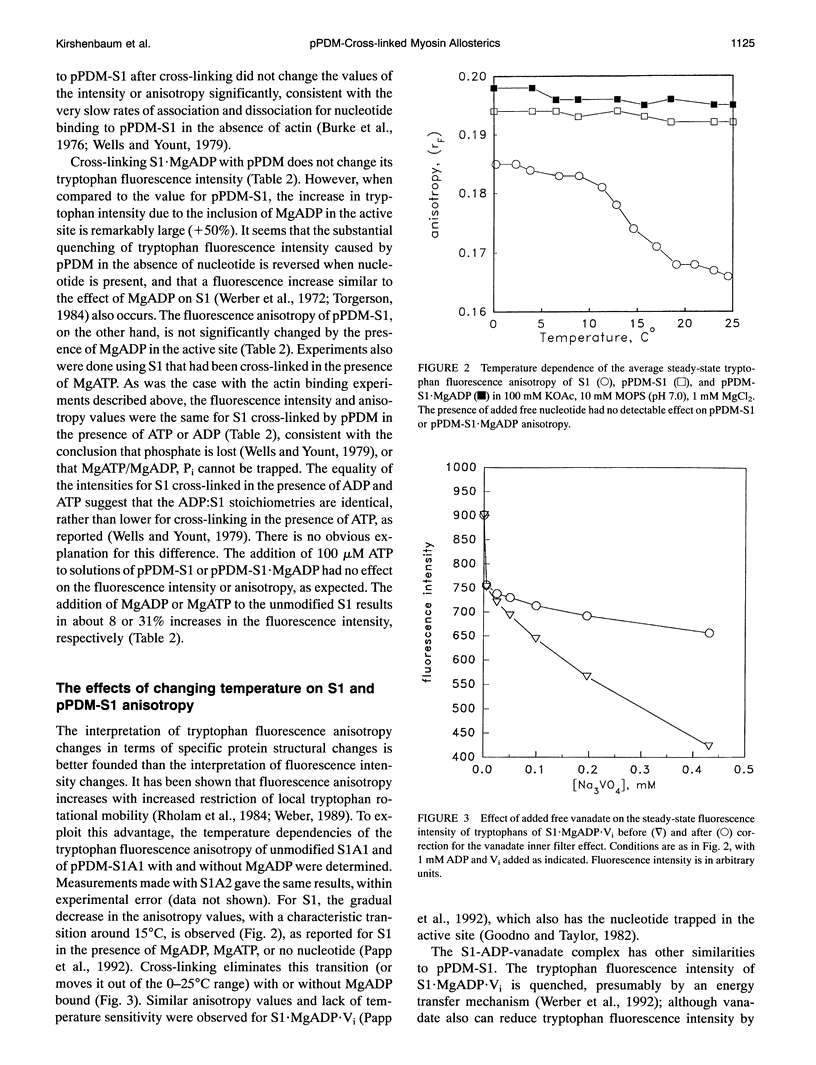




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aguirre R., Lin S. H., Gonsoulin F., Wang C. K., Cheung H. C. Characterization of the ethenoadenosine diphosphate binding site of myosin subfragment 1. Energetics of the equilibrium between two states of nucleotide.S1 and vanadate-induced global conformation changes detected by energy transfer. Biochemistry. 1989 Jan 24;28(2):799–807. doi: 10.1021/bi00428a058. [DOI] [PubMed] [Google Scholar]
- Ansari A., Jones C. M., Henry E. R., Hofrichter J., Eaton W. A. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992 Jun 26;256(5065):1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
- Audemard E., Bertrand R., Bonet A., Chaussepied P., Mornet D. Pathway for the communication between the ATPase and actin sites in myosin. J Muscle Res Cell Motil. 1988 Jun;9(3):197–218. doi: 10.1007/BF01773891. [DOI] [PubMed] [Google Scholar]
- Botts J., Takashi R., Torgerson P., Hozumi T., Muhlrad A., Mornet D., Morales M. F. On the mechanism of energy transduction in myosin subfragment 1. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2060–2064. doi: 10.1073/pnas.81.7.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botts J., Ue K., Hozumi T., Samet J. Consequences of reacting the thiols of myosin subfragment 1. Biochemistry. 1979 Nov 13;18(23):5157–5163. doi: 10.1021/bi00590a020. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burke M., Knight P. J. Studies on the role of sulfhydryls in the myosin ATPase. Characterization of the site of modification by the bifunctional sulfhydryl reagent p-N,N'-phenylenedimaleimide. J Biol Chem. 1980 Sep 25;255(18):8385–8387. [PubMed] [Google Scholar]
- Burke M., Reisler E. Effect of nucleotide binding on the proximity of the essential sulfhydryl groups of myosin. Chemical probing of movement of residues during conformational transitions. Biochemistry. 1977 Dec 13;16(25):5559–5563. doi: 10.1021/bi00644a026. [DOI] [PubMed] [Google Scholar]
- Burke M., Reisler F., Harrington W. F. Effect of bridging the two essential thiols of myosin on its spectral and actin-binding properties. Biochemistry. 1976 May 4;15(9):1923–1927. doi: 10.1021/bi00654a020. [DOI] [PubMed] [Google Scholar]
- Chalovich J. M., Greene L. E., Eisenberg E. Crosslinked myosin subfragment 1: a stable analogue of the subfragment-1.ATP complex. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4909–4913. doi: 10.1073/pnas.80.16.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantler P. D., Tao T., Stafford W. F., 3rd On the relationship between distance information derived from cross-linking and from resonance energy transfer, with specific reference to sites located on myosin heads. Biophys J. 1991 Jun;59(6):1242–1250. doi: 10.1016/S0006-3495(91)82339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase P. B., Beck T. W., Bursell J., Kushmerick M. J. Molecular charge dominates the inhibition of actomyosin in skinned muscle fibers by SH1 peptides. Biophys J. 1991 Aug;60(2):352–359. doi: 10.1016/S0006-3495(91)82060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussepied P., Morales M. F. Modifying preselected sites on proteins: the stretch of residues 633-642 of the myosin heavy chain is part of the actin-binding site. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7471–7475. doi: 10.1073/pnas.85.20.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung H. C., Gonsoulin F., Garland F. An investigation of the SH1-SH2 and SH1-ATPase distances in myosin subfragment-1 by resonance energy transfer using nanosecond fluorimetry. Biochim Biophys Acta. 1985 Nov 8;832(1):52–62. doi: 10.1016/0167-4838(85)90173-6. [DOI] [PubMed] [Google Scholar]
- Dalbey R. E., Weiel J., Yount R. G. Förster energy transfer measurements of thiol 1 to thiol 2 distances in myosin subfragment 1. Biochemistry. 1983 Sep 27;22(20):4696–4706. doi: 10.1021/bi00289a014. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Arata T. Effect of tryptic digestion of myosin subfragment-1 on its binding to F-actin. J Biochem. 1984 May;95(5):1343–1348. doi: 10.1093/oxfordjournals.jbchem.a134741. [DOI] [PubMed] [Google Scholar]
- Garland F., Gonsoulin F., Cheung H. C. The MgADP-induced decrease of the SH1-SH2 fluorescence resonance energy transfer distance of myosin subfragment 1 occurs in two kinetic steps. J Biol Chem. 1988 Aug 25;263(24):11621–11623. [PubMed] [Google Scholar]
- Goodno C. C., Taylor E. W. Inhibition of actomyosin ATPase by vanadate. Proc Natl Acad Sci U S A. 1982 Jan;79(1):21–25. doi: 10.1073/pnas.79.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene L. E., Chalovich J. M., Eisenberg E. Effect of nucleotide on the binding of N,N'-p-phenylenedimaleimide-modified S-1 to unregulated and regulated actin. Biochemistry. 1986 Feb 11;25(3):704–709. doi: 10.1021/bi00351a030. [DOI] [PubMed] [Google Scholar]
- Highsmith S., Eden D. Ligand-induced myosin subfragment 1 global conformational change. Biochemistry. 1990 May 1;29(17):4087–4093. doi: 10.1021/bi00469a010. [DOI] [PubMed] [Google Scholar]
- Highsmith S., Eden D. Myosin-ATP chemomechanics. Biochemistry. 1993 Mar 16;32(10):2455–2458. doi: 10.1021/bi00061a001. [DOI] [PubMed] [Google Scholar]
- Highsmith S. Electrostatic contributions to the binding of myosin and myosin-MgADP to F-actin in solution. Biochemistry. 1990 Nov 27;29(47):10690–10694. doi: 10.1021/bi00499a017. [DOI] [PubMed] [Google Scholar]
- Highsmith S. Interactions of the actin and nucleotide binding sites on myosin subfragment 1. J Biol Chem. 1976 Oct 25;251(20):6170–6172. [PubMed] [Google Scholar]
- Highsmith S., Kubinec M., Jaiswal D. K., Morimoto H., Williams P. G., Wemmer D. E. [2-3H]ATP synthesis and 3H NMR spectroscopy of enzyme-nucleotide complexes: ADP and ADP.Vi bound to myosin subfragment 1. J Biomol NMR. 1993 May;3(3):325–334. doi: 10.1007/BF00212518. [DOI] [PubMed] [Google Scholar]
- Highsmith S., Murphy A. J. Electrostatic changes at the actomyosin-subfragment 1 interface during force-generating reactions. Biochemistry. 1992 Jan 21;31(2):385–389. doi: 10.1021/bi00117a011. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T. Spatial proximity of ATP-sensitive tryptophanyl residue(s) and Cys-697 in myosin ATPase. J Biol Chem. 1992 Jul 25;267(21):14949–14954. [PubMed] [Google Scholar]
- Huston E. E., Grammer J. C., Yount R. G. Flexibility of the myosin heavy chain: direct evidence that the region containing SH1 and SH2 can move 10 A under the influence of nucleotide binding. Biochemistry. 1988 Dec 13;27(25):8945–8952. doi: 10.1021/bi00425a011. [DOI] [PubMed] [Google Scholar]
- Imamura K., Tada M., Tonomura Y. The pre-steady state of the myosin--adenosine triphosphate system. IV. Liberation of ADP from the myosin--ATP system and effects of modifiers on the phosphorylation of myosin. J Biochem. 1966 Mar;59(3):280–289. [PubMed] [Google Scholar]
- Johnson K. A., Taylor E. W. Intermediate states of subfragment 1 and actosubfragment 1 ATPase: reevaluation of the mechanism. Biochemistry. 1978 Aug 22;17(17):3432–3442. doi: 10.1021/bi00610a002. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Kasprzak A. A., Chaussepied P., Morales M. F. Location of a contact site between actin and myosin in the three-dimensional structure of the acto-S1 complex. Biochemistry. 1989 Nov 14;28(23):9230–9238. doi: 10.1021/bi00449a040. [DOI] [PubMed] [Google Scholar]
- Katoh T., Katoh H., Morita F. Actin-binding peptide obtained by the cyanogen bromide cleavage of the 20-kDa fragment of myosin subfragment-1. J Biol Chem. 1985 Jun 10;260(11):6723–6727. [PubMed] [Google Scholar]
- Katoh T., Morita F. Actin-binding peptides obtained from the C-terminal 24-kDa fragment of porcine aorta smooth muscle myosin subfragment-1 heavy chain. J Biol Chem. 1993 Feb 5;268(4):2380–2388. [PubMed] [Google Scholar]
- Lakowicz J. R., Freshwater G., Weber G. Nanosecond segmental mobilities of tryptophan residues in proteins observed by lifetime-resolved fluorescence anisotropies. Biophys J. 1980 Oct;32(1):591–601. doi: 10.1016/S0006-3495(80)84992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitsky D. I., Shnyrov V. L., Khvorov N. V., Bukatina A. E., Vedenkina N. S., Permyakov E. A., Nikolaeva O. P., Poglazov B. F. Effects of nucleotide binding on thermal transitions and domain structure of myosin subfragment 1. Eur J Biochem. 1992 Nov 1;209(3):829–835. doi: 10.1111/j.1432-1033.1992.tb17354.x. [DOI] [PubMed] [Google Scholar]
- Marston S., Weber A. The dissociation constant of the actin-heavy meromyosin subfragment-1 complex. Biochemistry. 1975 Aug 26;14(17):3868–3873. doi: 10.1021/bi00688a021. [DOI] [PubMed] [Google Scholar]
- Morita F., Ishigami F. Temperature dependence of the decay of the UV absorption difference spectrum of heavy meromyosin induced by adenosine triphosphate and inosine triphosphate. J Biochem. 1977 Feb;81(2):305–312. doi: 10.1093/oxfordjournals.jbchem.a131459. [DOI] [PubMed] [Google Scholar]
- Okamoto Y., Yount R. G. Identification of an active site peptide of skeletal myosin after photoaffinity labeling with N-(4-azido-2-nitrophenyl)-2-aminoethyl diphosphate. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1575–1579. doi: 10.1073/pnas.82.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp S., Eden D., Highsmith S. Nucleotide- and temperature-induced changes in myosin subfragment-1 structure. Biochim Biophys Acta. 1992 Oct 20;1159(3):267–273. doi: 10.1016/0167-4838(92)90055-i. [DOI] [PubMed] [Google Scholar]
- Reisler E., Burke M., Himmelfarb S., Harrington W. F. Spatial proximity of the two essential sulfhydryl groups of myosin. Biochemistry. 1974 Sep 10;13(19):3837–3840. doi: 10.1021/bi00716a001. [DOI] [PubMed] [Google Scholar]
- Spudich J. A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971 Aug 10;246(15):4866–4871. [PubMed] [Google Scholar]
- Sutoh K. Identification of myosin-binding sites on the actin sequence. Biochemistry. 1982 Jul 20;21(15):3654–3661. doi: 10.1021/bi00258a020. [DOI] [PubMed] [Google Scholar]
- Sutoh K. Mapping of actin-binding sites on the heavy chain of myosin subfragment 1. Biochemistry. 1983 Mar 29;22(7):1579–1585. doi: 10.1021/bi00276a009. [DOI] [PubMed] [Google Scholar]
- Sutoh K., Tokunaga M., Wakabayashi T. Electron microscopic mappings of myosin head with site-directed antibodies. J Mol Biol. 1989 Mar 20;206(2):357–363. doi: 10.1016/0022-2836(89)90485-3. [DOI] [PubMed] [Google Scholar]
- Suzuki R., Nishi N., Tokura S., Morita F. F-actin-binding synthetic heptapeptide having the amino acid sequence around the SH1 cysteinyl residue of myosin. J Biol Chem. 1987 Aug 25;262(24):11410–11412. [PubMed] [Google Scholar]
- Taylor E. W. Mechanism of actomyosin ATPase and the problem of muscle contraction. CRC Crit Rev Biochem. 1979;6(2):103–164. doi: 10.3109/10409237909102562. [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Sutoh K., Toyoshima C., Wakabayashi T. Location of the ATPase site of myosin determined by three-dimensional electron microscopy. Nature. 1987 Oct 15;329(6140):635–638. doi: 10.1038/329635a0. [DOI] [PubMed] [Google Scholar]
- Torgerson P. M. Tryptophan emission from myosin subfragment 1: acrylamide and nucleotide effect monitored by decay-associated spectra. Biochemistry. 1984 Jun 19;23(13):3002–3007. doi: 10.1021/bi00308a024. [DOI] [PubMed] [Google Scholar]
- Toyoshima Y. Y., Kron S. J., McNally E. M., Niebling K. R., Toyoshima C., Spudich J. A. Myosin subfragment-1 is sufficient to move actin filaments in vitro. Nature. 1987 Aug 6;328(6130):536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K., Tokunaga M., Kohno I., Sugimoto Y., Hamanaka T., Takezawa Y., Wakabayashi T., Amemiya Y. Small-angle synchrotron x-ray scattering reveals distinct shape changes of the myosin head during hydrolysis of ATP. Science. 1992 Oct 16;258(5081):443–447. doi: 10.1126/science.1411537. [DOI] [PubMed] [Google Scholar]
- Weeds A. G., Taylor R. S. Separation of subfragment-1 isoenzymes from rabbit skeletal muscle myosin. Nature. 1975 Sep 4;257(5521):54–56. doi: 10.1038/257054a0. [DOI] [PubMed] [Google Scholar]
- Wells J. A., Yount R. G. Active site trapping of nucleotides by crosslinking two sulfhydryls in myosin subfragment 1. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4966–4970. doi: 10.1073/pnas.76.10.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werber M. M., Peyser Y. M., Muhlrad A. Characterization of stable beryllium fluoride, aluminum fluoride, and vanadate containing myosin subfragment 1-nucleotide complexes. Biochemistry. 1992 Aug 11;31(31):7190–7197. doi: 10.1021/bi00146a023. [DOI] [PubMed] [Google Scholar]
- Werber M. M., Szent-Györgyi A. G., Fasman G. D. Fluorescence studies on heavy meromyosin-substrate interaction. Biochemistry. 1972 Jul 18;11(15):2872–2883. doi: 10.1021/bi00765a021. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. Binding manner of actin to the lysine-rich sequence of myosin subfragment 1 in the presence and absence of ATP. Biochemistry. 1989 Jun 27;28(13):5573–5577. doi: 10.1021/bi00439a035. [DOI] [PubMed] [Google Scholar]



