Abstract
The distribution of 20 variable regions resulting from insertion-deletion events in the genomes of the tubercle bacilli has been evaluated in a total of 100 strains of Mycobacterium tuberculosis, Mycobacterium africanum, Mycobacterium canettii, Mycobacterium microti, and Mycobacterium bovis. This approach showed that the majority of these polymorphisms did not occur independently in the different strains of the M. tuberculosis complex but, rather, resulted from ancient, irreversible genetic events in common progenitor strains. Based on the presence or absence of an M. tuberculosis specific deletion (TbD1), M. tuberculosis strains can be divided into ancestral and “modern” strains, the latter comprising representatives of major epidemics like the Beijing, Haarlem, and African M. tuberculosis clusters. Furthermore, successive loss of DNA, reflected by region of difference 9 and other subsequent deletions, was identified for an evolutionary lineage represented by M. africanum, M. microti, and M. bovis that diverged from the progenitor of the present M. tuberculosis strains before TbD1 occurred. These findings contradict the often-presented hypothesis that M. tuberculosis, the etiological agent of human tuberculosis evolved from M. bovis, the agent of bovine disease. M. canettii and ancestral M. tuberculosis strains lack none of these deleted regions, and, therefore, seem to be direct descendants of tubercle bacilli that existed before the M. africanum→M. bovis lineage separated from the M. tuberculosis lineage. This observation suggests that the common ancestor of the tubercle bacilli resembled M. tuberculosis or M. canettii and could well have been a human pathogen already.
Keywords: evolution‖diagnostic‖identification
The mycobacteria grouped in the Mycobacterium tuberculosis complex are characterized by 99.9% similarity at the nucleotide level and identical 16S rRNA sequences (1, 2) but differ widely in terms of their host tropisms, phenotypes, and pathogenicity. Assuming that they all are derived from a common ancestor, it is intriguing that some are exclusively human (M. tuberculosis, Mycobacterium africanum, Mycobacterium canettii) or rodent pathogens (Mycobacterium microti), whereas others have a wide host spectrum (Mycobacterium bovis). What was the genetic organization of the last common ancestor of the tubercle bacilli, and in which host did it live? Which genetic events may have contributed to the fact that the host spectrum is so different and often specific? Where and when did M. tuberculosis evolve? Answers to these questions are important for a better understanding of the pathogenicity and the global epidemiology of tuberculosis and may help to anticipate future trends in the spread of the disease.
Because of the unusually high degree of conservation in their housekeeping genes, it has been suggested that the members of the M. tuberculosis complex underwent an evolutionary bottleneck at the time of speciation, estimated to have occurred roughly 15,000–20,000 years ago (2). Also, it has been speculated that M. tuberculosis, the most widespread etiological agent of human tuberculosis has evolved from M. bovis, the agent of bovine tuberculosis, by specific adaptation of an animal pathogen to the human host (3). However, both hypotheses were proposed before the whole genome sequence of M. tuberculosis (4) was available and before comparative genomics uncovered several variable genomic regions in the members of the M. tuberculosis complex. Differential hybridization arrays identified 14 regions of difference (RD1–14), ranging in size from 2 to 12.7 kb, that were absent from bacillus Calmette–Guérin Pasteur relative to M. tuberculosis H37Rv (5, 6). In parallel, six regions, H37Rv related deletions (RvD)1–5, and M. tuberculosis specific deletion 1 (TbD1), that were absent from the M. tuberculosis H37Rv genome relative to other members of the M. tuberculosis complex were revealed by comparative genomics approaches employing pulsed-field gel electrophoresis techniques (5, 7) and in silico comparisons of the near complete M. bovis AF2122/97 genome sequence and the M. tuberculosis H37Rv sequence.
In the present study, we have analyzed the distribution of these 20 variable regions situated around the genome (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org) in a representative and diverse set of 100 strains belonging to the M. tuberculosis complex. The strains were isolated from different hosts and from a broad range of geographic origins, and exhibit a wide spectrum of typing characteristics like IS6110 and spoligotype hybridization patterns or variable-number tandem repeats of mycobacterial interspersed repetitive units (MIRU-VNTR; refs. 8 and 9). We have found striking evidence that deletion of certain variable genomic regions did not occur independently in the different strains of the M. tuberculosis complex; assuming that there is little or no recombination of chromosomal segments between the various lineages of the complex, this allows us to propose a completely new scenario for the evolution of the M. tuberculosis complex and the origin of human tuberculosis.
Material and Methods
Bacterial Strains.
The 100 M. tuberculosis complex strains were composed of 46 M. tuberculosis strains isolated in 30 countries, 14 M. africanum strains, 28 M. bovis strains originating in 5 countries, 2 M. bovis bacillus Calmette–Guérin vaccine strains (Pasteur and Japan), 5 M. microti strains, and 5 M. canettii strains. The strains were isolated from human and animal sources and were selected to represent a wide diversity, including 60 strains that have been used in a multicenter study (8). The M. africanum strains were retrieved from the collection of the Wadsworth Center (New York State Department of Health, Albany, NY), whereas the majority of the M. bovis isolates came from the collection of the University of Zaragoza (Zaragoza, Spain). Four M. canettii strains are from the culture collection of the Pasteur Institute (Paris, France). The strains have been extensively characterized by reference typing methods, i.e., IS6110 restriction fragment length polymorphism (RFLP) typing and spoligotyping. M. tuberculosis H37Rv, M. tuberculosis H37Ra, M. tuberculosis CDC1551, M. bovis AF2122/97, M. microti OV254, and M. canettii CIPT 140010059 were included as reference strains. DNA was prepared as described (10).
Genome Comparisons and Primer Design.
For preliminary genome comparisons between M. tuberculosis and M. bovis, web sites http://genolist.pasteur.fr/TubercuList/and http://www.sanger.ac.uk/Projects/M_bovis/, as well as in-house databases, were used. For primer design, sequences inside or flanking RD and RvD regions were obtained from the same web sites. Primers were designed by using the Primer3 web site http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi that would amplify ≈500 bp fragments in the reference strains (Table 1).
RD-PCR Analysis.
Reactions were performed in 96-well plates and contained per reaction 1.25 μl of 10× PCR buffer (600 mM Tris⋅HCl, pH 8.8/20 mM MgCl2/170 mM (NH4)2SO4/100 mM β-mercaptoethanol), 1.25 μl 20 mM nucleotide mix, 50 nM of each primer, 1–10 ng of template DNA, 10% DMSO, 0.2 units Taq polymerase (GIBCO-BRL), and sterile distilled water to 12.5 μl. Thermal cycling was performed on a PTC-100 amplifier (MJ Research, Cambridge, MA) with an initial denaturation step of 90 s at 95°C, followed by 35 cycles of 30 s at 95°C, 1 min at 58°C, and 4 min at 72°C.
Sequencing of katG, gyrA, oxyR, pncA, mmpl6 Genes and RD, TbD1 Junction Regions.
PCR products were obtained as described above, using the primers listed in Table 1.
For primer elimination, 6 μl of PCR product was incubated with 1 unit of shrimp alkaline phosphatase (United States Biochemical), 10 units of exonuclease I (United States Biochemical), and 2 μl of 5 × buffer (200 mM Tris⋅HCl, pH 8.8/5 mM MgCl2) for 15 min at 37°C and then for 15 min at 80°C. To this reaction mixture, 2 μl of Big Dye sequencing mix (Applied Biosystems), 2 μl (2 μM) of primer, and 3 μl of 5 × buffer (5 mM MgCl2/200 mM Tris⋅HCl, pH 8.8) were added, and 35 cycles (96°C for 30 sec, 56°C for 15 sec, and 60°C for 4 min) were performed in a thermocycler (MJ Research, Cambridge, MA). DNA was precipitated by using 80 μl of 76% ethanol, centrifuged, rinsed with 70% ethanol, and dried. Reactions were dissolved in 2 μl of formamide/EDTA buffer, denatured and loaded onto 48-cm 4% polyacrylamide gels, and electrophoresis was performed on 377 automated DNA sequencers (Applied Biosystems) for 10 to 12 h. Alternatively, reactions were dissolved in 0.3 mM EDTA buffer and subjected to automated sequencing on a Model 3700 DNA sequencer (Applied Biosystems). Reactions generally gave between 500–700 bp of unambiguous sequence.
Accession Numbers.
The sequence of the TbD1 region from the ancestral M. tuberculosis strain no. 74 (8) containing genes mmpS6 and mmpL6 was deposited in the EMBL database under accession no. AJ426486. Sequences bordering RD4, RD7, RD8, RD9, and RD10 in bacillus Calmette–Guérin were deposited in the EMBL database under accession nos. AJ003103, AJ007301, AJ131210, Y18604, and AJ132559, respectively.
Results
Variable Genomic Regions and Their Occurrence in the Members of the M. tuberculosis Complex.
The PCR screening assay for the 20 variable regions (Table 1) within 46 M. tuberculosis, 14 M. africanum, 5 M. canettii, 5 M. microti, 28 M. bovis, and 2 bacillus Calmette–Guérin strains used oligonucleotides internal to known RDs and RvDs, as well as oligonucleotides flanking these regions (Table 1). This approach generated a large data set that was robust, highly reliable, and internally controlled, because PCR amplicons obtained with the internal primer pair correlated with the absence of an appropriately sized amplicon with the flanking primer-pair, and vice versa.
According to the conservation of junction sequences flanking the variable regions, three types of regions were distinguished, each one having a different importance as an evolutionary marker. The first type included mobile genetic elements, like the prophages phiRv1 (RD3) and phiRv2 (RD11) and insertion sequences IS1532 (RD6) and IS6110 (RD5), whose distribution in the tubercle bacilli was highly divergent (see Table 2, which is published as supporting information on the PNAS web site). The second type of deletion is mediated by homologous recombination between adjacent IS6110 insertion elements resulting in the loss of the intervening DNA segment (RvD2, RvD3, RvD4, and RvD5; ref. 7) and is variable from strain to strain (Table 2).
The third type includes deletions whose bordering genomic regions typically do not contain repetitive sequences. Often, this type of deletion occurred in coding regions resulting in the truncation of genes that are still intact in other strains of the M. tuberculosis complex. The exact mechanism leading to this type of deletion remains obscure, but possibly rare strand slippage errors of DNA polymerase may have contributed to this event. As shown in detail below, RD1, RD2, RD4, RD7, RD8, RD9, RD10, RD12, RD13, RD14, and TbD1 are representatives of this third group whose distribution among the 100 strains allows us to propose an evolutionary scenario for the members of the M. tuberculosis complex that identifies M. tuberculosis and/or M. canettii as most closely related to the common ancestor of the tubercle bacilli.
M. tuberculosis Strains.
Investigation of the 46 M. tuberculosis strains by deletion analysis revealed that most RD regions were present in all M. tuberculosis strains tested (Table 2). Only regions RD3 and RD11, corresponding to the two prophages phiRv1 and phiRv2 of M. tuberculosis H37Rv (4), RD6, containing the insertion sequence IS1532, and RD5, that is flanked by a copy of IS6110 (5), were absent in some strains. This observation is an important one, as it implies that M. tuberculosis strains are highly conserved with respect to RD1, RD2, RD4, RD7, RD8, RD9, RD10, RD12, RD13, and RD14, and that these RDs represent regions that can differentiate M. tuberculosis strains independently of their geographical origin and their typing characteristics from certain other members of the M. tuberculosis complex. Furthermore, this finding suggests that these regions may be involved in the host specificity of M. tuberculosis.
In contrast, the presence or absence of RvD regions in M. tuberculosis strains was variable. The region that showed the greatest variability was RvD2, because 18 of 46 tested M. tuberculosis strains did not carry the RvD2 region. Strains with a high copy number of IS6110 (>14) missed regions RvD2 to RvD5 more often than strains with only a few copies. As an example, all six tested strains belonging to the Beijing cluster (8) lacked regions RvD2 and RvD3. This finding is in agreement with the proposed involvement of recombination of two adjacent copies of IS6110 in this deletion event (7).
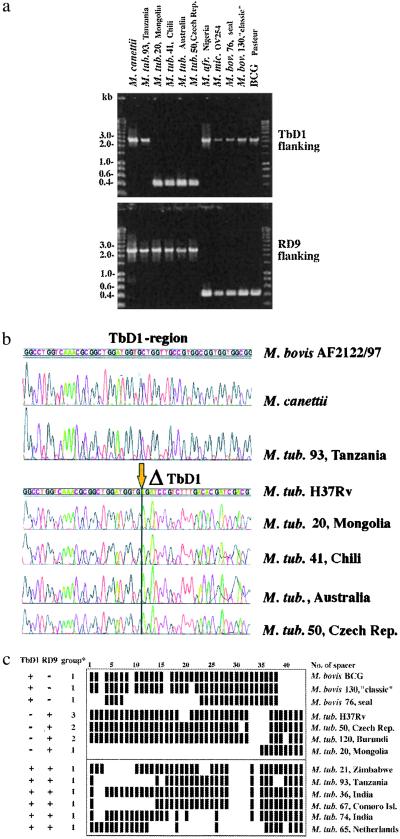
However, the most surprising finding concerning the RvD regions was that TbD1 was absent from 40 of the tested M. tuberculosis strains (87%), including representative strains from major epidemics such as the Haarlem, Beijing, and Africa clusters (8). To accentuate this result, we named this region “M. tuberculosis specific deletion 1” (TbD1). In silico sequence comparison of M. tuberculosis H37Rv with the corresponding section in M. bovis AF2122/97 revealed that, in M. bovis, this locus comprises two genes encoding membrane proteins belonging to a large family, whereas in M. tuberculosis H37Rv, one of these genes (mmpS6) was absent and the second was truncated (mmpL6). Unlike the RvD2-RvD5 deletions, the TbD1 region is not flanked by a copy of IS6110 in M. tuberculosis H37Rv, suggesting that insertion elements were not involved in the deletion of the 2,153-bp fragment. To investigate further whether the 40 M. tuberculosis strains lacking the TbD1 region had the same genomic organization of this locus as M. tuberculosis H37Rv, we amplified the TbD1-junction regions of the various strains by PCR using primers flanking the deleted region (Table 1). This approach showed that the size of the amplicons obtained from multiple strains was uniform (Fig. 1A), and subsequent sequence analysis of the PCR products revealed that in all tested TbD1-deleted strains the sequence of the junction regions was identical to that of M. tuberculosis H37Rv (Fig. 1B). The perfect conservation of the junction sequences in TbD1-deleted strains of wide geographical diversity suggests that the genetic event which resulted in the deletion occurred in a common progenitor. However, six M. tuberculosis strains, all characterized by very few or no copies of IS6110, and spoligotypes that resembled each other (Fig. 1C), still had the TbD1 region present. Interestingly, these six strains also were clustered together by MIRU-VNTR analysis (9).
Figure 1.
(A) Amplicons obtained from strains that have the indicated genomic region present or deleted. Sizes of amplicons in each group are uniform. Numbers correspond to strain designation used in refs. 8 and 9. (B) Sequences in the TbD1 region obtained from strains of various geographic regions. Numbers correspond to strain designation used in refs. 8 and 9. (C) Spoligotypes of selected M. tuberculosis and M. bovis strains. Numbers correspond to strain designation used in refs. 8 and 9. *, groups based on katGc463/gyrAc95 sequence polymorphism defined by Sreevatsan et al. (2).
Analysis of partial gene sequences of oxyR, pncA, katG, and gyrA which have been described as variable between different tubercle bacilli (2, 11–13) revealed that all tested M. tuberculosis strains showed oxyR and pncA partial sequences typical for M. tuberculosis [oxyR, nucleotide 285 (oxyR285):G; pncA, codon 57 (pncA57):CAC]. Based on the katG codon 463 (katG463) and gyrA codon 95 (gyrA95) sequence polymorphism, Sreevatsan et al. (2) defined three groups among the tubercle bacilli: group 1 showed katG463 CTG (Leu), gyrA95 ACC (Thr); group 2 exhibited katG463 CGG (Arg), gyrA95 ACC (Thr); and group 3 showed katG463 CGG (Arg), gyrA95 AGC (Ser). According to this scheme, 16 of the 46 tested M. tuberculosis strains in our study belonged to group 1, whereas 27 strains belonged to group 2, and only 3 isolates belonged to group 3. From the 40 strains that were deleted for region TbD1, 9 showed characteristics of group 1, including the strains belonging to the Beijing cluster, 28 showed characteristics of group 2, including the strains from the Haarlem and Africa clusters, and 3 showed characteristics of group 3, including H37Rv and H37Ra. Most interestingly, all six M. tuberculosis strains where the TbD1 region was not deleted, contained a leucine (CTG) at katG463, which was described as characteristic for ancestral M. tuberculosis strains (group 1; ref. 2). As shown in Fig. 2, this finding suggests that during the evolution of M. tuberculosis, the katG mutation at codon 463 CTG (Leu) → CGG (Arg) occurred in a progenitor strain that had region TbD1 deleted. This proposal is supported by the finding that strains belonging to group 1 may or may not have deleted region TbD1, whereas all 30 strains belonging to groups 2 and 3 lacked TbD1 (Fig. 2). Furthermore, all strains of groups 2 and 3 characteristically lacked spacer sequences 33–36 in the direct repeat (DR) region (Fig. 1C). It appears that such spacers may be lost but not gained (14). Therefore, TbD1-deleted strains will be referred to hereafter as “modern” M. tuberculosis strains.
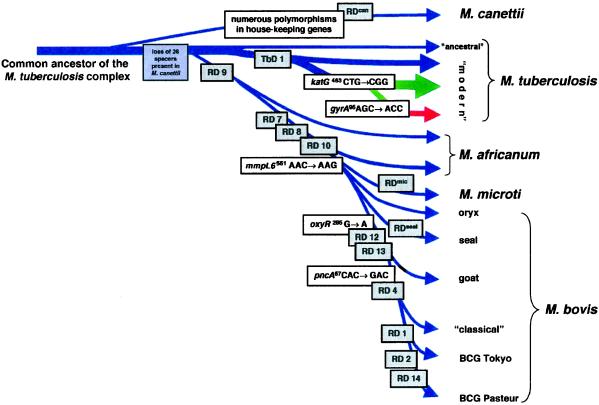
Figure 2.
Scheme of the proposed evolutionary pathway of the tubercle bacilli illustrating successive loss of DNA in certain lineages (gray boxes). The scheme is based on the presence or absence of conserved deleted regions and on sequence polymorphisms in five selected genes. Note that the distances between certain branches may not correspond to actual phylogenetic differences calculated by other methods. Blue arrows indicate that strains are characterized by katG463. CTG (Leu), gyrA95 ACC (Thr), typical for group 1 organisms. Green arrows indicate that strains belong to group 2 characterized by katG463 CGG (Arg), gyrA95 ACC (Thr). The red arrow indicates that strains belong to group 3, characterized by katG463 CGG (Arg), gyrA95 AGC (Ser), as defined by Sreevatsan et al. (2).
M. canettii.
M. canettii is a very rare, smooth variant of M. tuberculosis and is usually isolated from patients from, or with connection to, Africa. Although it shares identical 16S rRNA sequences with the other members of the M. tuberculosis complex, M. canettii strains differ in many respects, including polymorphisms in certain house-keeping genes, IS1081 copy number, colony morphology, and the lipid content of the cell wall (15, 16). Therefore, we were surprised to find that in M. canettii, all of the RD, RvD, and TbD1 regions except the prophages (phiRv1, phiRv2) were present. In contrast, we identified a region (RDcan) being specifically absent from all five M. canettii strains that partially overlapped RD12 (Fig. 2).
The conservation of the RD, RvD, and TbD1 regions in the genome of M. canettii in conjunction with the many described and observed differences suggest that M. canettii diverged from the common ancestor of the M. tuberculosis complex before RD, RvD, and TbD1 occurred in the lineages of tubercle bacilli (Fig. 2). This hypothesis is supported by the finding that M. canettii was shown to carry 26 unique spacer sequences in the DR region (14) that are no longer present in any other member of the M. tuberculosis complex. Therefore, M. canettii represents a fascinating tubercle bacillus, whose detailed genomic analysis may reveal further insights into the evolution of the M. tuberculosis complex.
M. africanum.
The isolates designated as M. africanum studied here originate from West and East African sources. Eleven strains were isolated in Sierra Leone, Nigeria, and Guinea, and 2 strains were isolated in Uganda. One strain comes from the Netherlands.
For the 11 West African isolates, RD analysis indicated that these strains all lack the RD9 region containing cobL. Sequence analysis of the RD9 junction region showed that the genetic organization of this locus in West African strains was identical to that of M. bovis and M. microti in that the 5′ part of cobL as well as the genes Rv2073c and Rv2074c were absent. In addition, six strains (two from Sierra Leone and four from Guinea) also lacked RD7, RD8, and RD10 (Table 2). The junction sequences bordering RD7, RD8, and RD10, like those for RD9, were identical to those of M. bovis and M. microti strains. As regards the two prophages phiRv1 and phiRv2, the West African strains all contained phiRv2, whereas phiRv1 was absent. No variability was seen for the RvD regions. RvD1-RvD5 and TbD1 were present in all tested West African strains. This result shows that M. africanum prevalent in West Africa can be differentiated from modern M. tuberculosis by at least two variable genetic markers, namely the absence of region RD9 and the presence of region TbD1.
In contrast, for East African M. africanum and for the isolate from the Netherlands, no genetic marker was found which could differentiate them from M. tuberculosis strains. With the exception of prophage phiRv1 (RD3), the three strains from Uganda and the Netherlands did not exhibit any of the RD deletions, but lacked the TbD1 region, as do modern M. tuberculosis strains. The absence of the TbD1 region also was confirmed by sequence analysis of the TbD1 junction region, which was found to be identical to that of TbD1-deleted M. tuberculosis strains. These results indicate a very close genetic relationship of these strains to M. tuberculosis and suggest that they should be regarded as M. tuberculosis rather than M. africanum strains.
M. microti.
M. microti strains were isolated in the 1930s from voles (17) and, more recently, from immuno-suppressed patients (18). These strains are characterized by an identical, characteristic spoligotype, but differ in their IS6110 profiles. Both the vole and the human isolates lacked regions RD7, RD8, RD9, and RD10, as well as a region that is specifically deleted from M. microti (RDmic). RDmic was revealed by a detailed comparative genomics study of M. microti isolates (P. Brodin, personal communication) using clones from an M. microti bacterial artificial chromosome library. RDmic partially overlaps RD1 from bacillus Calmette–Guérin (data not shown). Furthermore, vole isolates missed part of the RD5 region, whereas this region was present in the human isolate. As the junction region of RD5 in M. microti was different from that in bacillus Calmette–Guérin (data not shown), RD5 was not used as an evolutionary marker.
M. bovis and M. bovis Bacillus Calmette–Guérin.
M. bovis has a very large host spectrum infecting many mammalian species, including man. The collection of M. bovis strains that were screened for the RD and RvD regions consisted of 2 bacillus Calmette–Guérin strains and 18 “classical” M. bovis strains that were generally characterized by only one or two copies of IS6110 from bovine, llama, and human sources, in addition to three goat isolates, three seal isolates, two oryx isolates, and two M. bovis strains from humans that presented a higher number of IS6110 copies.
Excluding prophages, the distribution of RDs allowed us to differentiate five main groups among the tested M. bovis strains. The first group was formed by strains that lack RD7, RD8, RD9, and RD10. Representatives of this group are three seal isolates and two human isolates containing between three and five copies of IS6110 (data not shown). Two oryx isolates harboring between 17 and 20 copies of IS6110 formed the second group that lacked parts of RD5 in addition to RD7-RD10 and very closely resembled the M. microti isolates. However, they did not show RDmic, the deletion characteristic of M. microti strains (data not shown). Analysis of partial oxyR and pncA sequences from strains belonging to groups one and two showed sequence polymorphisms characteristic of M. tuberculosis strains (oxyR285:G, pncA57:CAC; refs. 12 and 13).
Group three consists of goat isolates that lack regions RD5, RD7, RD8, RD9, RD10, RD12, and RD13. As described by Aranaz et al. (20), these strains exhibited an adenosine at position 285 of the oxyR pseudogene that is specific for classical M. bovis strains, whereas the sequence of the pncA57 polymorphism was identical to that in M. tuberculosis. This finding is in good agreement with our results from sequence analysis (Table 2) and the finding that, except for RD4, the goat isolates displayed the same deletions as classical M. bovis strains.
Taken together, this suggests that the oxyR285 mutation (G → A) occurred in M. bovis strains before RD4 was lost. Interestingly, the most common M. bovis strains—classical M. bovis (21), isolated from cattle from Argentina, the Netherlands, the UK and Spain, as well as from humans (e.g., multidrug resistant M. bovis from Spain; ref. 22)—showed the greatest number of RD deletions and seem to have undergone the greatest loss of DNA relative to other members of the M. tuberculosis complex. These lacked regions RD4, RD5, RD6, RD7, RD8, RD9, RD10, RD12, and RD13, confirming results obtained with reference strains (5, 6). These strains, together with the two bacillus Calmette–Guérin strains, were the only ones that showed the pncA57 polymorphism GAC (Asp) in addition to the oxyR285 mutation (G → A) characteristic of M. bovis. Analysis of bacillus Calmette–Guérin strains indicate that bacillus Calmette–Guérin lacked the same RD regions as classical M. bovis strains, in addition to RD1, RD2, and RD14, which apparently occurred during and after the attenuation process (Fig. 2; refs. 6 and 23).
In contrast to RDs, the RvD regions were highly conserved in the M. bovis strains. With the exception of the two IS6110-rich oryx isolates that lacked RvD2, RvD3, and RvD4, all other strains had the five RvD regions present. It is particularly noteworthy that TbD1 was present in all M. bovis strains.
However, except for the two human isolates containing between three and five copies of IS6110 from group 1, strains designated as M. bovis showed a single nucleotide polymorphism in the TbD1 region at codon 551 (AAG) of the mmpL6 gene relative to M. canettii, M. africanum, and ancestral M. tuberculosis strains, which are characterized by codon AAC. Even the strains isolated from seals and from oryx with oxyR or pncA loci like those of M. tuberculosis, and with fewer deleted regions than the classical M. bovis strains, showed the mmpL6551AAG polymorphism typical for M. bovis and M. microti (Table 2, Fig. 2). As such, this polymorphism could serve as a very useful genetic marker for the differentiation of strains that lack RD7, RD8, RD9, and RD10 and have been classified as M. bovis or M. africanum but may differ from other strains of the same taxon.
Discussion
Origin of Human Tuberculosis.
For many years, it was thought that human tuberculosis evolved from the bovine disease by adaptation of an animal pathogen to the human host (3). This hypothesis is based on the property of M. tuberculosis to be almost exclusively a human pathogen, whereas M. bovis has a much broader host range. However, the results from this study unambiguously show that M. bovis has undergone numerous deletions relative to M. tuberculosis. This finding is confirmed by the preliminary analysis of the near complete genome sequence of M. bovis AF2122/97, a classical M. bovis strain isolated from cattle, which revealed no new gene clusters that were confined specifically to M. bovis. This result indicates that the genome of M. bovis is smaller than that of M. tuberculosis (24). It seems plausible that M. bovis is the final member of a separate lineage represented by M. africanum (RD9), M. microti (RD7, RD8, RD9, RD10) and M. bovis (RD4, RD5, RD7, RD8, RD9, RD10, RD12, RD13; ref. 25) that branched from the progenitor of M. tuberculosis isolates. Successive loss of DNA may have contributed to clonal expansion and the appearance of more successful pathogens in certain new hosts.
Whether the progenitor of extant M. tuberculosis strains was already a human pathogen when the M. africanum → M. bovis lineage separated from the M. tuberculosis lineage is a subject for speculation. However, we have two reasons to believe that this was the case. First, the six ancestral M. tuberculosis strains (TbD1+, RD9+; Fig. 1C) that resemble the last common ancestor before the separation of M. tuberculosis and M. africanum are all human pathogens. Second, M. canettii, which probably diverged from the common ancestor of today's M. tuberculosis strains before any other known member of the M. tuberculosis complex, is also a human pathogen. Taken together, this means that those tubercle bacilli, which are thought to most closely resemble the progenitor of M. tuberculosis, are human and not animal pathogens. It is also intriguing that most of these strains were of African or Indian origin (Fig. 1C). It is likely that these ancestral strains predominantly originated from endemic foci (15, 26), whereas modern M. tuberculosis strains that have lost TbD1 may represent epidemic M. tuberculosis strains that were introduced into the same geographical regions more recently as a consequence of the worldwide spread of the tuberculosis epidemic.
The Evolutionary Time Scale of the M. tuberculosis Complex.
Because of the high sequence conservation in housekeeping genes, Sreevatsan et al. (2) hypothesized that the tubercle bacilli encountered a major bottleneck 15,000–20,000 years ago. As the conservation of the TbD1 junction sequence in all tested TbD1-deleted strains suggests a descent from a single clone, the TbD1 deletion is a perfect indicator that modern M. tuberculosis strains that account for the vast majority of today's tuberculosis cases definitely underwent such a bottleneck and then spread around the world.
As described in detail in Results, our analysis showed that the katG(463) CTG → CGG and the subsequent gyrA(95) ACC → AGC mutations that were used by Sreevatsan et al. to designate groups 2 and 3 of their proposed evolutionary pathway of the tubercle bacilli (2) occurred in a lineage of M. tuberculosis strains that had already lost TbD1 (Fig. 2). Although deletions are more stable markers than point mutations, which may be subject to reversion, a perfect correlation of deletion and point mutation data were found for the tested strains.
This information, together with results from a recent study by Fletcher et al. (27), who have shown that M. tuberculosis DNAs amplified from naturally mummified Hungarian villagers from the 18th and 19th century belonged to katG463/gyrA95 groups 2 and 3, suggests that the TbD1 deletion occurred in the lineage of M. tuberculosis before the 18th century. This could mean that the dramatic increase of tuberculosis cases later in the 18th century in Europe mainly involved modern M. tuberculosis strains. In addition, it shows that tuberculosis was caused by M. tuberculosis and not by M. bovis, a fact which is also described for cases in rural medieval England (28).
There is good evidence that mycobacterial infections occurred in man several thousand years ago. We know that tuberculosis occurred in Egypt during the reign of the pharaohs because spinal and rib lesions pathognomonic of tuberculosis have been identified in mummies from that period (29). Identification of acid-fast bacilli, as well as PCR amplification of IS6110 from Peruvian mummies (30), also suggest that tuberculosis existed in preColumbian societies of Central and South America. To estimate when the TbD1 bottleneck occurred, it would be helpful to know whether the Egyptian and South American mummies carried M. tuberculosis DNA that did or did not have TbD1 deleted.
The other major bottleneck, which seems to have occurred for members of the M. africanum → M. microti → M. bovis lineage, is reflected by RD9 and the subsequent RD7, RD8, and RD10 deletions (Fig. 2). These deletions seem to have occurred in the progenitor of tubercle bacilli that—today—show natural host spectra as diverse as humans in Africa, voles on the Orkney Isles (UK), seals in Argentina, goats in Spain, and badgers in the UK. For this reason, it is difficult to imagine that the spread and adaptation of RD9-deleted bacteria to their specific hosts could have appeared within the postulated 15,000–20,000 years of speciation of the M. tuberculosis complex.
However, more insight into this matter could be gained by RD analysis of ancient DNA samples, e.g., mycobacterial DNA isolated from a 17,000-year-old bison skeleton (19). The Mycobacterium whose DNA was amplified showed a spoligotype that was most closely related to patterns of M. africanum and could have been an early representative of the lineage M. africanum → M. bovis. With the TbD1 and RD9 junction sequences that we supply here, PCR analyses of ancient DNAs should enable very focused studies to be undertaken to learn more about the time scale within which the members of the M. tuberculosis complex have evolved.
Concluding Comments.
Our study provides an overview of the diversity and conservation of variable regions in a broad range of tubercle bacilli. Deletion analysis of 100 strains from various hosts and countries has identified some evolutionarily “old” M. canettii, M. tuberculosis, and M. africanum strains, most of them of African origin, as well as modern M. tuberculosis strains, the latter including representatives from major epidemic clusters like Beijing, Haarlem, and Africa. The use of deletion analysis in conjunction with molecular typing and analysis of specific mutations was shown to represent a very powerful approach for the study of the evolution of the tubercle bacilli and for the identification of evolutionary markers. In a more practical perspective, these regions, primarily RD9 and TbD1 but also RD1, RD2, RD4, RD7, RD8, RD10, RD12, and RD13 represent very interesting candidates for the development of powerful diagnostic tools for the rapid and unambiguous identification of members of the M. tuberculosis complex. This genetic approach for differentiation can now be used to replace the often confusing traditional division of the M. tuberculosis complex into rigidly defined subspecies.
Moreover, functional analyses will show whether the TbD1 deletion confers some selective advantage to modern M. tuberculosis, or whether other circumstances contributed to the pandemic of the TbD1-deleted M. tuberculosis strains.
Supplementary Material
Acknowledgments
We thank Philip Supply for his encouragement and help to initiate this study. We also thank Carlos Martin, Tim Stinear, and Veronique Vincent for fruitful discussions. This work was supported by the Institut Pasteur (PTR 2000/35), the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses, the European Union (QLK2-CT-1999-01093, QLRT-CT-2000-00630), the Department of Environment, Food and Rural Affairs (GB), the Association Française Raoul Follereau, and The Wellcome Trust.
Abbreviations
- TbD1
M. tuberculosis specific deletion 1
- RvD
H37Rv related deletion
- RD
region of difference
Footnotes
References
- 1.Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger E C. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stead W W, Eisenach K D, Cave M D, Beggs M L, Templeton G L, Thoen C O, Bates J H. Am J Respir Crit Care Med. 1995;151:1267–1268. doi: 10.1164/ajrccm/151.4.1267. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole S T. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 6.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 7.Brosch R, Philipp W J, Stavropoulos E, Colston M J, Cole S T, Gordon S V. Infect Immun. 1999;67:5768–5774. doi: 10.1128/iai.67.11.5768-5774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L W, Yakrus M A, et al. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. J Clin Microbiol. 2001;39:3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Soolingen D, de Haas P E W, Hermans P W M, van Embden J D A. Methods Enzymol. 1994;235:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- 11.Heym B, Honore N, Truffot-Pernot C, Banerjee A, Schurra C, Jacobs W R, Jr, van Embden J D, Grosset J H, Cole S T. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 12.Scorpio A, Collins D, Whipple D, Cave D, Bates J, Zhang Y. J Clin Microbiol. 1997;35:106–110. doi: 10.1128/jcm.35.1.106-110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sreevatsan S, Escalante P, Pan X, Gillies D A, 2nd, Siddiqui S, Khalaf C N, Kreiswirth B N, Bifani P, Adams L G, Ficht T, et al. J Clin Microbiol. 1996;34:2007–2010. doi: 10.1128/jcm.34.8.2007-2010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Embden J D, van Gorkom T, Kremer K, Jansen R, van Der Zeijst B A, Schouls L M. J Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Soolingen D, Hoogenboezem T, de Haas P E, Hermans P W, Koedam M A, Teppema K S, Brennan P J, Besra G S, Portaels F, Top J, et al. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 16.Papa F, Laszlo A, David H L, Daffe M. Acta Leprol. 1989;7, Suppl. 1:98–101. [PubMed] [Google Scholar]
- 17.Wells A Q, Suppl . Lancet. 1937. , 1221. [Google Scholar]
- 18.van Soolingen D, van der Zanden A G, de Haas P E, Noordhoek G T, Kiers A, Foudraine N A, Portaels F, Kolk A H, Kremer K, van Embden J D. J Clin Microbiol. 1998;36:1840–1845. doi: 10.1128/jcm.36.7.1840-1845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothschild B M, Martin L D, Lev G, Bercovier H, Bar-Gal G K, Greenblatt C, Donoghue H, Spigelman M, Brittain D. Clin Infect Dis. 2001;33:305–311. doi: 10.1086/321886. [DOI] [PubMed] [Google Scholar]
- 20.Aranaz A, Liebana E, Gomez-Mampaso E, Galan J C, Cousins D, Ortega A, Blazquez J, Baquero F, Mateos A, Suarez G, Dominguez L. Int J Syst Bacteriol. 1999;49:1263–1273. doi: 10.1099/00207713-49-3-1263. [DOI] [PubMed] [Google Scholar]
- 21.van Soolingen D, de Haas P E W, Haagsma J, Eger T, Hermans P W M, Ritacco V, Alito A, van Embden J D A. J Clin Microbiol. 1994;32:2425–2433. doi: 10.1128/jcm.32.10.2425-2433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samper S, Martin C, Pinedo A, Rivero A, Blazquez J, Baquero F, van Soolingen D, van Embden J. Aids. 1997;11:1237–1242. doi: 10.1097/00002030-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 23.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon S V, Eiglmeier K, Garnier T, Brosch R, Parkhill J, Barrell B, Cole S T, Hewinson R G. Tuberculosis. 2001;81:157–163. doi: 10.1054/tube.2000.0269. [DOI] [PubMed] [Google Scholar]
- 25.Brosch R, Gordon S V, Eiglmeier K, Garnier T, Tekaia F, Yeramanian E, Cole S T. In: Molecular Genetics of Mycobacteria. Hatful G F, Jacobs W R Jr, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 19–36. [Google Scholar]
- 26.Radhakrishnan I, K, M Y, Kumar R A, Mundayoor S. J Clin Microbiol. 2001;39:1683. doi: 10.1128/JCM.39.4.1683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fletcher, H. A., Donoghue, H. D., Holton, J., Pap, I. & Spigelman, M. (2002) Am. J. Phys. Anthropol., in press. [DOI] [PubMed]
- 28.Mays S, Taylor G M, Legge A J, Young D B, Turner-Walker G. Am J Phys Anthropol. 2001;114:298–311. doi: 10.1002/ajpa.1042. [DOI] [PubMed] [Google Scholar]
- 29.Nerlich A G, Haas C J, Zink A, Szeimies U, Hagedorn H G. Lancet. 1997;350:1404. doi: 10.1016/S0140-6736(05)65185-9. [DOI] [PubMed] [Google Scholar]
- 30.Salo W L, Aufderheide A C, Buikstra J, Holcomb T A. Proc Natl Acad Sci USA. 1994;91:2091–2094. doi: 10.1073/pnas.91.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.