Abstract
The somatic accumulation of defective mitochondria causes human degenerative syndromes, senescence in fungi, and male sterility in plants. These diverse phenomena may result from conflicts between natural selection at different levels of organization. Such conflicts are fundamental to the evolution of cooperating groups, from cells to populations. We present a model in which defective mitochondrial genomes accumulate because of a within-cell replication advantage when among-cell selection for efficient respiration is relaxed. We tested the model by using experimental populations of the yeast Saccharomyces cerevisiae. We constructed yeast strains that were heteroplasmic for mitochondrial mutations that destroy the ability to respire (the petite phenotype) and followed the accumulation of mitochondrial defects in cultures with different effective population sizes. As predicted by the model, the inability to respire evolved only in small populations of S. cerevisiae, where among-cell selection favoring cells that can respire was reduced relative to within-cell selection favoring parasitic mitochondria. In a control experiment, mitochondrial point mutations that confer resistance to chloramphenicol showed no tendency to change in frequency under any culture conditions. The accumulation of some mitochondrial defects is therefore an evolutionary process, involving multiple levels of selection. The relative intensities of within- and among-cell selection may also explain the tissue specificity of human mitochondrial defects.
Mitochondrial disorders are recognized as the cause of an increasingly large number of progressive genetic diseases in humans, including Leber's hereditary optic neuropathy, Kearns–Sayre syndrome, cancer, and senescence (1). In many of these disorders, the degeneration of specific tissues (especially those with high respiratory demands) is accompanied by the accumulation of mitochondria carrying inactivating deletions or point mutations. There is increasing evidence that the frequency of mutant mitochondria within heteroplasmic cells increases with age (2) and across successive generations within a maternal lineage (3). Distinct syndromes are caused by different frequencies of mitochondrial mutants within tissues or by the accumulation of the same deletion in different tissues. Mitochondrial disorders are not unique to humans. In plants, mtDNA rearrangements can cause male sterility, a trait that has been put to widespread use in the production of hybrid seed (4). In some fungi, plasmids formed from mtDNA increase in copy number during somatic growth, resulting in senescence (5).
A common explanation for the accumulation of mitochondrial defects is that the reactive byproducts of oxidative respiration cause high mutation rates in mtDNA. Mitochondria also lack some of the DNA repair pathways that operate in the nucleus to reduce mutation rates (6). However, some defects may accumulate because mutant mitochondrial chromosomes have a fitness advantage over functional mitochondrial genomes within cells (7–14). The opportunity for natural selection to occur within cells stems from the fact that each cell contains multiple mitochondrial genomes that replicate independently of the cell. Selection among the replicating genomes may favor fast-replicating variants, even if they are deleterious to the cell in which they reside. The mutant mitochondria may be held in check by natural selection among cells, or among whole organisms, favoring efficient respiration. Syndromes caused by the accumulation of mutant mitochondria would then be expected to arise when selection among cells, tissues, or organisms is relaxed and mitochondrial evolution is driven by selection within cells. This model of parasitic vs. altruistic mitochondrial genomes is analogous to other classic examples of conflicting levels of selection, such as the evolution of cooperative (or altruistic) social behaviors that reduce the fitness of individuals within groups relative to selfishness, but which increase the relative fitness of cooperative social groups.
Budding yeast (Saccharomyces cerevisiae) are ideally suited to experimental testing of these models, particularly as they apply to mitochondrial chromosomes within cells. From the perspective of the mitochondrial genome, altruistic behavior takes the form of encoding, transcribing, and translating the mitochondrial gene products necessary for respiration even if that reduces the rate or efficiency of its own replication. Selfish or parasitic mitochondrial genomes are those that are replicated or transmitted more efficiently than the wild type but do not encode respiratory functions. Unlike most eukaryotes, yeast can meet their energetic needs by fermentation alone, with the result that nonrespiring mutants are viable and readily studied. At a frequency near 1% for most strains, yeast cells spontaneously become fixed, or homoplasmic, for mitochondrial genomes from which most of the chromosome has been deleted (15, 16). The remaining fragment becomes tandemly repeated to yield a final chromosome size near the wild-type 80 kb. Because the repeat unit typically contains an origin of replication, it has been hypothesized that such genomes may enjoy a replication advantage over wild-type mtDNA (15, 17). Yeast cells homoplasmic for such mtDNA deletions show the petite phenotype, which is diagnosed by the inability to grow on carbon sources such as glycerol that require respiration, and by the formation of smaller colonies on fermentable carbon sources such as dextrose. Petite genomes face an among-cell selective disadvantage of about 30% when competing against isogenic wild-type cells on minimal dextrose media (18). The deletion of mitochondrial genes abolishes the mitochondrial transcription and translation machinery required for respiration, thus there is little variation among different petites in cellular fitness.
However, petite genomes vary significantly in their rates of mitotic and meiotic transmission. Some petites, like typical point mutations in mitochondrial genes, segregate from heteroplasmic cells (those with a mixture of genetically distinct mitochondrial genomes) at rates equal to wild-type mitochondrial genomes. Other petites are preferentially inherited from heteroplasmic parental cells so that some daughter cells are fixed for the mutant mitochondrial genome and show the petite phenotype. The fraction of mitotic progeny of a petite x wild-type cross that is fixed for the petite mitochondrial genome is referred to as the suppressivity of that petite. Hypersuppressive petites completely exclude wild types from meiotic and mitotic progeny, and therefore represent the extreme in parasitic mitochondrial genomes. Recent evidence indicates that preferential replication of hypersuppressive petite chromosomes, rather than biased segregation during cell budding, excludes wild-type mitochondrial genomes (19). However, petites lacking origins of replication are stably maintained in mitotically reproducing cultures (20), apparently by a different replication mechanism than that involved in the replication advantage of hypersuppressives (19).
We use a mathematical model to identify the parameters that influence the relative importance of within- vs. among-cell selection on mitochondrial mutations. In experimental yeast populations we vary the intensity of among-cell selection by varying the effective population size. We show that mitochondrial mutations in S. cerevisiae are favored by natural selection within cells, but are consistently kept at very low frequencies in large populations because natural selection among cells favors wild-type mitochondria. The evolution of mitochondrial genomes within cells is a uniquely powerful example of multilevel selection (cf. ref. 21), because antagonism among levels of selection can be explicitly demonstrated and experimentally manipulated, and because the conflict among levels of selection is relevant to medically important phenomena.
Methods
Simulation Model.
To illustrate how multilevel selection models can be applied to mitochondrial genomes within cells, we developed a simulation model. The model belongs to a general class of multilevel selection models where the population is subdivided into discrete groups of individuals, and selection occurs among groups as well as among individuals within groups (22–25). In our case, the discrete groups are cells that contain numerous individual mitochondrial genomes. The specific model is very similar to Haldane's (26) model of selection for altruistic behavior within and among human tribes [also applied to budding viscous populations such as insect colonies (27)], except that we make assumptions specific to groups of mitochondrial genomes within asexual yeast populations. For example, all individuals (mitochondrial genomes) that colonize new groups (cells) are selected from only a single group from the previous generation (the parent cell)—the so-called “propagule pool” model (28). We also assumed there was no migration of individuals among established groups.
We assumed a constant number (N) of asexually reproducing cells. Within the ith parent cell, there is a frequency of mutant mitochondrial genomes (pi) that are distributed randomly across mitochondrial compartments within the cell. Daughter cells inherit the average frequency of mutant mitochondrial genomes from their parent. However, we assumed that founder effects at each round of replication or genetic drift during cell growth would cause the effective population size of mitochondrial genomes (M) to be relatively small (29). Genetic analyses of the rate at which mitochondrial genotypes segregate from heteroplasmic yeast cells show that despite the relatively large number of mitochondria per cell (≈100), the effective population size of mitochondrial genomes is on the order of 2–5 per cell (29). We therefore assumed the number of defective mitochondrial genomes per daughter cell had binomial distribution with a mean of Mpi, which generated among-cell variation in the frequency of mutant mitochondria (30). Within each daughter cell, defective mitochondrial genomes enjoyed a replication advantage of (1 + σ) relative to wild-type mitochondrial genomes. After t generations of within-cell evolution, where t is the number of rounds of mitochondrial genome replication per cell generation, each cell was assigned a fitness (w) proportional to the new frequency of functional mitochondria within the cell (pi′). Specifically, w = 1 − γpi′, where γ is the severity of the effect of defective mitochondria on cell fitness (30). Cells in the next generation were made by selecting N parents (with replacement) from among the cells of the previous generation, and with a probability proportional to the their fitness. The source code (in C) is available from the authors on request.
Experimental Populations of Saccharomyces.
To test the simulation model experimentally, we used laboratory populations of S. cerevisiae. Each population was founded by one of five isogenic ancestral strains that were heteroplasmic for an independently isolated spontaneous mitochondrial deletion. Mitochondrial petite mutants were obtained from a haploid strain and crossed to an isogenic haploid to construct the diploid, heteroplasmic ancestors of experimental populations. Among the parameters identified by the simulations as being particularly important, the size of the cell population is the one that can be most precisely manipulated. From each ancestor we established five replicate yeast populations. These 25 populations were then propagated through 150 generations at small, medium, and large population sizes (effective population sizes were approximately 10, 250, and 18,000, respectively) to determine the effect of population size on the fates of mutant mitochondrial genomes.
Spontaneously arising petite strains were isolated from a derivative of strain S288c (MATa, leu2Δ URA3; ATCC catalog no. 200876) by streaking cells on yeast extract/peptone/dextrose agar (1% yeast extract/1% peptone/2% dextrose) and picking isolated colonies that were substantially smaller than the rest. From these candidates, genotypes that could not respire were identified by streaking onto yeast extract/peptone/glycerol (YPGlyc) agar (1% yeast extract/1% peptone/3% glycerol). Petites are unable to grow on glycerol because it cannot be fermented and therefore requires respiratory competence. Petites were crossed with an isogenic haploid strain (MATα, LEU2 ura3Δ), and the diploids were induced to undergo meiosis and sporulation to confirm mitochondrial inheritance of the inability to respire. The resulting asci were dissected, and tetrads were replica-plated to agar media lacking uracil or leucine (to confirm Mendelian 2:2 segregation of the known nuclear LEU2 and URA3 markers) and to YPGlyc (to test for 4:0 wild-type:petite segregation, indicating that the petite phenotype resulted from a nonsuppressive mitochondrial mutation).
For the first five mitochondrial petites identified in this way, the diploid strain produced for the test cross was the founder of a set of five small, five medium, and five large replicate experimental populations. Medium and large populations were propagated at a constant 30° in 13 × 100 mm borosilicate tubes containing 0.2 ml of yeast extract/peptone/dextrose (YPD) with the dextrose concentration reduced to 0.2%. At 24-h intervals, large populations were vortexed, and 2 μl, containing on average 6,000 cells, was transferred to fresh sterile medium. Medium populations were diluted 1,666-fold in sterile water before transfer, reducing the average number of cells transferred to 30. Small populations were propagated on YPD agar by randomly choosing a single colony of each population (representing the descendants of a single cell) after 48 h of growth and streaking it onto a fresh plate. Effective population sizes, estimated as the harmonic mean of population sizes at hourly intervals throughout the growth cycle, were 1.8 × 104, 250, and 10 for large, medium, and small populations, respectively. Frequencies of petite cells in each population were determined after 150 cell generations by plating a suspension of cells diluted so as to give 100–200 colonies per plate onto YPD agar. After 2 days' growth they were replica-plated to yeast extract/peptone/glycerol, and the numbers of petite and respiration-competent colonies were counted. Based on the simulation results, we predicted greater frequencies of petite cells in the smaller populations.
For the control experiment, spontaneous chloramphenicol-resistant mutants were isolated on yeast extract/peptone/glycerol agar containing 0.4% chloramphenicol. Genetic confirmation of mitochondrial inheritance, construction of heteroplasmic diploids, propagation of experimental populations, and estimation of chloramphenicol-resistant cell frequencies were then performed exactly as for the petite experiment. Like the ability to respire, chloramphenicol resistance is expressed in heteroplasmic cells. During the propagation of the small populations, 2 of the 25 populations (1 replicate each founded by 2 of the 5 ancestors) became petite. It was not possible to determine whether the petite mitochondrial genotypes fixed in these two populations were derived from chloramphenicol-resistant or wild-type mitochondria. Chloramphenicol resistance is diagnosed as growth on nonfermentable substrates in the presence of chloramphenicol (an inhibitor of translation in wild-type mitochondria), but a petite cell cannot grow on such media even if its mitochondrial chromosomes were derived from resistant chromosomes. These two small populations were therefore not counted in determining the final frequencies of chloramphenicol resistance.
Results and Discussion
That selection occurs at multiple levels of organization is fundamental to the evolution of cooperating units at many levels of biological organization—chromosomes within genomes, organelles within cells, cells within bodies, and individuals within social groups. One of the first formal models of this process was by Haldane (26), who supposed that when humans are clustered into cooperative tribes, behaviors that reduce individual fitness might nevertheless spread if they benefit the group. The evolution of “altruistic” behavior therefore depends on populations being structured such that selection among cooperative groups is at least as important as selection among individuals within those groups (23–25). For example, when groups are comprised of few individuals, genetic drift and founder events tend to diminish the within-group variance that drives the selection favoring selfish behavior within groups and enhance the among-group variance that drives the selection favoring altruism among groups (24, 25). It is now generally recognized that many phenomena are influenced by multiple levels of selection; these include cooperative behavior in social insects and mole rats (31), the evolution of reduced virulence in some pathogens (32), the female-biased sex ratio in fig wasps and spiders (33, 34), and numerous examples of selfish genetic elements (35). In fact, the emergence of selection at increasing levels of organization is often regarded as the driving force behind the major transitions in the evolution of life on earth, such as the evolution of genomes of cooperative genes, bodies of cooperating cells, and societies of cooperating individuals (36).
Our simulation shows that the evolution of mitochondrial disorders can be treated as a classical multilevel selection problem. It also identifies the specific properties of cells, and mitochondrial dynamics within cells, that control the accumulation of mitochondrial defects. In our situation, functional mitochondrial genomes are altruistic; they are selected against within the cell because they contribute to the respiration machinery of the cell at the expense of their own rate of replication. In the simulation, defective mitochondria either accumulated to fixation within cells or were completely lost, and this outcome depended predictably on the relative importance of within-cell vs. among-cell selection. For example, when the population size of mitochondrial genomes within cells was small, within-cell genetic variance was diminished and among-cell genetic variance was enhanced. This among-cell variance increased the relative importance of among-cell selection (29) and prevented the accumulation of defective mitochondrial genomes. Defective mitochondrial genomes were also less likely to accumulate when cell proliferation was rapid relative to the rate of replication of mtDNA replication within cells.
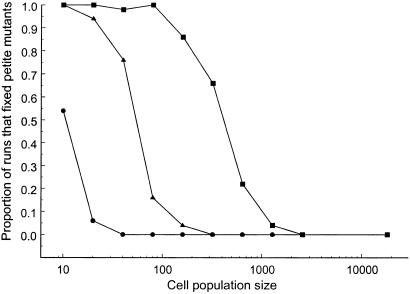
We focused our experimental work on manipulating the size of the cell population. Population size is a major determinant of the efficiency of selection in any population. As population size decreases, fitness differences among genotypes are increasingly outweighed by sampling error, and genotype frequencies are increasingly determined by genetic drift instead of selection. This effect is reflected in our model by greater fixation probabilities of defective mitochondria when cell populations were small and among-cell selection was weak (Fig. 1).
Figure 1.
Mitochondrial defects are fixed more frequently in model runs with smaller cell populations. Proportion of simulation runs that resulted in the accumulation of mitochondrial defects are shown as a function of the cell population size, N. The different symbols represent different within-cell replication advantages for the mitochondrial defects: (●) σ = 0.10, (▴) σ = 0.15, (■) σ = 0.20. Fifty replicates were run for each parameter value. The other model parameters were γ = 1.0, t = 5, and M = 15 (see text for details).
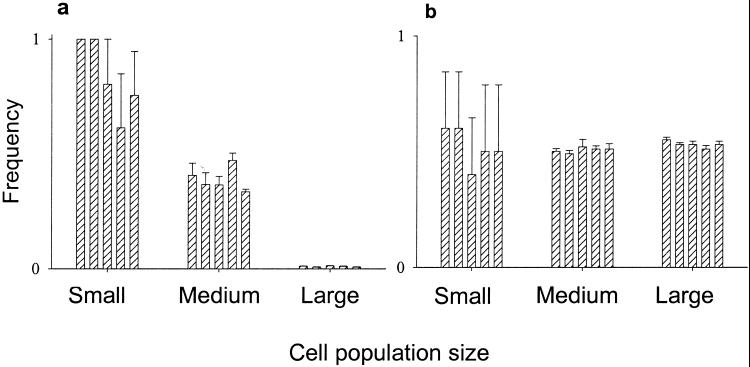
The accumulation of mitochondrial defects in experimental populations of S. cerevisiae fits the predictions of the multilevel selection model. There was a strong and inverse relationship between population size and the success of mutant mitochondrial genomes (Fig. 2a). The efficient among-cell selection occurring in the large populations reduced the final frequency of petites to less than 1% in all replicates, whereas 20 of the 25 small populations became entirely petite. Results for the medium population sizes were intermediate, with frequencies of petite cells ranging from 0.33 to 0.47. The effect of population size was highly significant (ANOVA, P < 0.001), and there were no significant differences among the populations descended from different heteroplasmic ancestors (P = 0.493) nor among replicate populations founded by the same heteroplasmic ancestor (P = 0.549).
Figure 2.
Frequencies after 150 generations of yeast cells (a) unable to respire (petite) and (b) resistant to chloramphenicol. Each bar represents the mean of five replicate populations founded by a diploid heteroplasmic ancestor, except that frequencies for two of the small population chloramphenicol treatment are based on four replicates (see Methods). Error bars represent SEMs. The same five ancestors were the founders of small, medium, and large populations.
As a control, we repeated this experiment by using populations that initially were heteroplasmic for a point mutation conferring resistance to chloramphenicol. Unlike petites, these mutations would be expected to confer neither a within-cell replication advantage nor a large among-cell disadvantage, thus their average frequencies should be unaffected by population size. This prediction was supported: final frequencies of chloramphenicol-resistant cells were near 0.5 in all replicates of large and medium populations founded by cells heteroplasmic for independent chloramphenicol-resistance mutations (Fig. 2b). Small populations quickly became fixed for either resistant or sensitive cells, at nearly equal frequencies.
The chloramphenicol control experiment also eliminates the spread of new mutations as the explanation for the petite frequencies in small populations. Among the 25 small populations propagated in the chloramphenicol control, only 2 became petite. This result indicates that the rate at which new deletions arise, although appreciable, is insufficient to explain the high frequencies with which small populations became petite. However, we note that intracellular selection and high mutation rates are not mutually exclusive mechanisms for mitochondrial syndromes, and a combination of the two processes may operate in some cell or tissue types.
We have shown that in S. cerevisiae, within-cell selection favors the accumulation of mitochondrial defects (petite mutants), with functional mitochondria being maintained only when among-cell selection is relatively intense. The specific mechanism by which mitochondrial defects realize a within-cell advantage is not clear; it could involve mitochondrial over-replication per se (15) or differences in how the proliferation (5) or degradation (37) of defective mitochondria are regulated by the nucleus. In fact, nuclear genes or nuclear–cytoplasmic interactions are known to influence the severity of several mitochondrial disorders (1), and a nuclear gene involved in the transmission advantage of some mutant mitochondria has been identified in yeast (38). Regardless of the specific mechanism of within-cell fitness advantage, this is a promising experimental system for understanding dynamics of mitochondrial defects within and among cells, and more generally, the conditions under which selection at higher levels of organization subsumes, or is undermined by, selection among entities at lower levels of organization.
Several aspects of human mitochondrial pathologies are consistent with the existence of conflicting levels of selection, especially the somatic accumulation of mitochondrial defects during aging (2) and across generations within a maternal lineage (3), as well as the simultaneous accumulation of the same mitochondrial mutation at different rates in different tissues. A within-cell advantage for mutant mitochondrial genomes has been suggested by the results of experiments with mice (11) and with cultures of human cells (7, 9, 10, 13). In some experiments, this within-cell advantage has been tissue-specific, appearing in one tissue or cell type but being absent or reversed in another (11). This pattern has been interpreted as an indication that the replication of mtDNA is regulated by one or more factors encoded by the nucleus that are expressed in a tissue-specific way and that discriminate among genetically different mitochondrial chromosomes (13). Alternatively, our model predicts that such tissue-specific differences may result from differences in the general properties of cells (14), or even the culture conditions, that would alter the relative intensities of within- and among-cell selection. We expect mitochondrial deletions to proliferate most rapidly in tissues with low cell turnover (which diminishes among-cell selection) or with rapid rates of mtDNA replication (which enhances within-cell selection). In fact, human mitochondrial defects accumulate most rapidly in high energy-demanding tissue (e.g., muscle) or in tissues with relatively low cell turnover (e.g., nerves) (11). Approaching these disorders as an evolutionary problem involving multiple levels of selection may lead to a broader understanding of the origins of mitochondrial defects and the evolutionary dynamics of organelle genomes.
Acknowledgments
We thank J. Antonovics, S. Church, J. Curran, P. Ingvarsson, M. Kidwell, D. McCauley, M. Silman, and two anonymous reviewers for discussions and comments on the manuscript. This work was supported by National Science Foundation Grants DEB-9876397 and DEB-0078546 (to D.R.T.), an Alfred P. Sloan Foundation grant and National Science Foundation Grant DEB-0075594 (to C.W.Z.), and by a Wake Forest University Research Fellowship (to E.C.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wallace DC. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 2.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 3.Smith K H, Johns D R, Heher K L, Miller N R. Arch Ophthalmol. 1993;111:1486–1490. doi: 10.1001/archopht.1993.01090110052022. [DOI] [PubMed] [Google Scholar]
- 4.Schnabel P S, Wise R P. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- 5.Bertrand H. Annu Rev Phytopathol. 2000;38:397–422. doi: 10.1146/annurev.phyto.38.1.397. [DOI] [PubMed] [Google Scholar]
- 6.Clayton D A, Doda J N, Friedberg E C. Proc Natl Acad Sci USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneda M, Chomyn A, Martinuzzi A, Hurko O, Attardi G. Proc Natl Acad Sci USA. 1992;89:11164–11168. doi: 10.1073/pnas.89.23.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spelbrink J N, Zwart R, Van Galen M J M, Van den Bogert C. Curr Genet. 1997;32:115–124. doi: 10.1007/s002940050255. [DOI] [PubMed] [Google Scholar]
- 9.Holt I J, Dunbar D R, Jacobs H T. Hum Mol Genet. 1997;6:1251–1260. doi: 10.1093/hmg/6.8.1251. [DOI] [PubMed] [Google Scholar]
- 10.Dunbar D R, Moonie P A, Jacobs H T, Holt I J. Proc Natl Acad Sci USA. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenuth J P, Peterson A C, Shoubridge E A. Nat Genet. 1997;16:93. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 12.Chinnery P F, Thorburn D R, Samuels D C, White S L, Dahl H-H M, Turnbull D M, Lightowlers R N, Howell N. Trends Genet. 2000;16:500–505. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- 13.Moraes C T, Kenyon L, Hao H. Mol Biol Cell. 1999;10:3345–3356. doi: 10.1091/mbc.10.10.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofhaus G, Gatterman N. Biol Chem. 1999;380:871–877. doi: 10.1515/BC.1999.107. [DOI] [PubMed] [Google Scholar]
- 15.Gingold E B. In: The Division and Segregation of Organelles. Boffey S A, Lloyd D, editors. New York: Cambridge Univ. Press; 1988. pp. 149–170. [Google Scholar]
- 16.Dujon B. In: The Molecular Biology of the Yeast Saccharomyces. Strathern J N, Jones E W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1981. pp. 505–635. [Google Scholar]
- 17.de Zamaroczy M, Marotta R, Faugeron-Fony G, Goursot R, Mangin M, Baldacci G, Bernardi G. Nature (London) 1981;292:75–78. doi: 10.1038/292075a0. [DOI] [PubMed] [Google Scholar]
- 18.Zeyl C, de Visser J A G M. Genetics. 2001;157:53–61. doi: 10.1093/genetics/157.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacAlpine D M, Kolesar J, Okamoto K, Butow R A, Perlman P S. EMBO J. 2001;20:1807–1817. doi: 10.1093/emboj/20.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fangman W L, Henly J W, Churchill G, Brewer B J. Mol Cell Biol. 1989;9:1917–1921. doi: 10.1128/mcb.9.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodnight C, Stevens L. Am Nat. 1997;150:S59–S79. doi: 10.1086/286050. [DOI] [PubMed] [Google Scholar]
- 22.Maynard Smith J. Nature (London) 1964;201:1145–1146. [Google Scholar]
- 23.Price G. Nature (London) 1970;277:520–521. doi: 10.1038/227520a0. [DOI] [PubMed] [Google Scholar]
- 24.Wilson D S. Am Nat. 1977;111:157–185. [Google Scholar]
- 25.Wade M. Am Nat. 1985;125:61–73. [Google Scholar]
- 26.Haldane J B S. The Causes of Evolution. London: Greens; 1932. [Google Scholar]
- 27.Goodnight K F. Am Nat. 1992;140:1028–1040. doi: 10.1086/285454. [DOI] [PubMed] [Google Scholar]
- 28.Wade M. Quart Rev Biol. 1978;53:101–114. [Google Scholar]
- 29.Birky C W., Jr Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- 30.Bergstrom C, Pritchard J. Genetics. 1998;149:2135–2146. doi: 10.1093/genetics/149.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman P, Jarvis J, Alexander R. The Biology of the Naked Mole Rat. Princeton: Princeton Univ. Press; 1991. [Google Scholar]
- 32.Herre E. Science. 1993;259:1442–1446. doi: 10.1126/science.259.5100.1442. [DOI] [PubMed] [Google Scholar]
- 33.Colwell R. Nature (London) 1981;190:401–404. [Google Scholar]
- 34.Aviles L. Am Nat. 1993;142:320–345. [Google Scholar]
- 35.Hurst L, Atlan A, Bengtsson B. Quart Rev Biol. 1996;71:317–364. doi: 10.1086/419442. [DOI] [PubMed] [Google Scholar]
- 36.Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. New York: Freeman; 1995. [Google Scholar]
- 37.Gray A. BioEssays. 1997;19:161–166. [Google Scholar]
- 38.Zweifel S G, Fangman W L. Genetics. 1991;128:241–249. doi: 10.1093/genetics/128.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]