Abstract
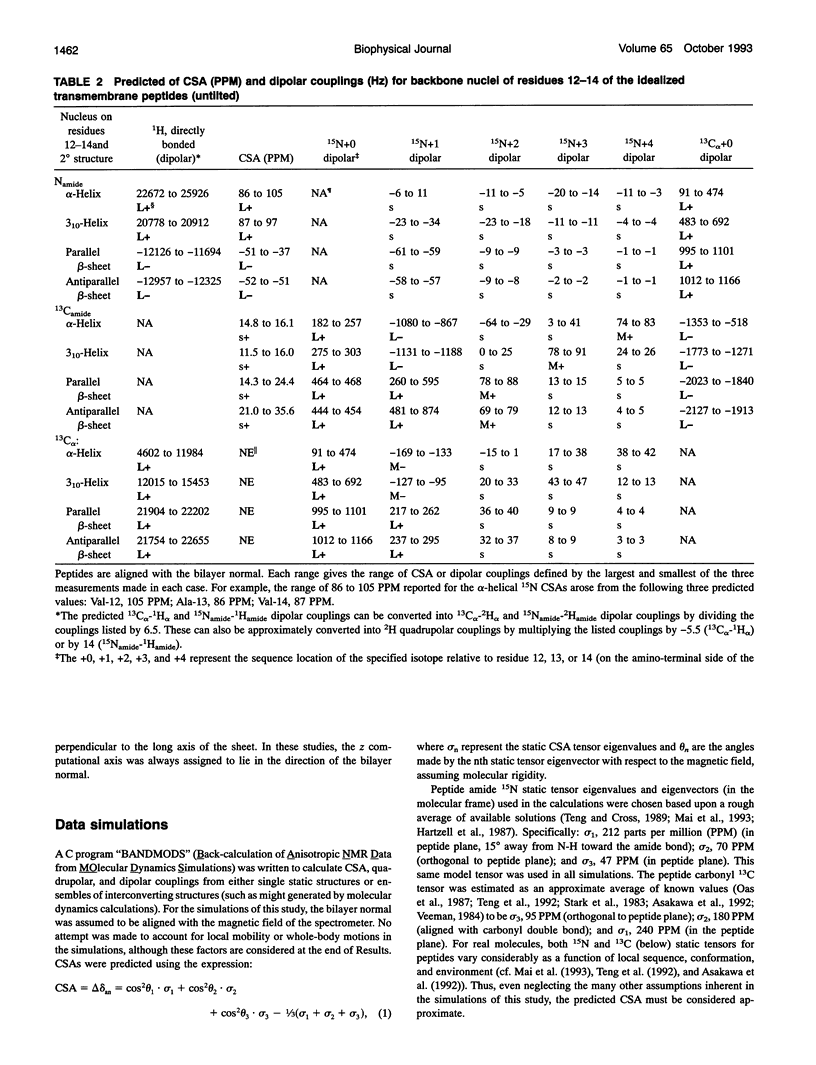
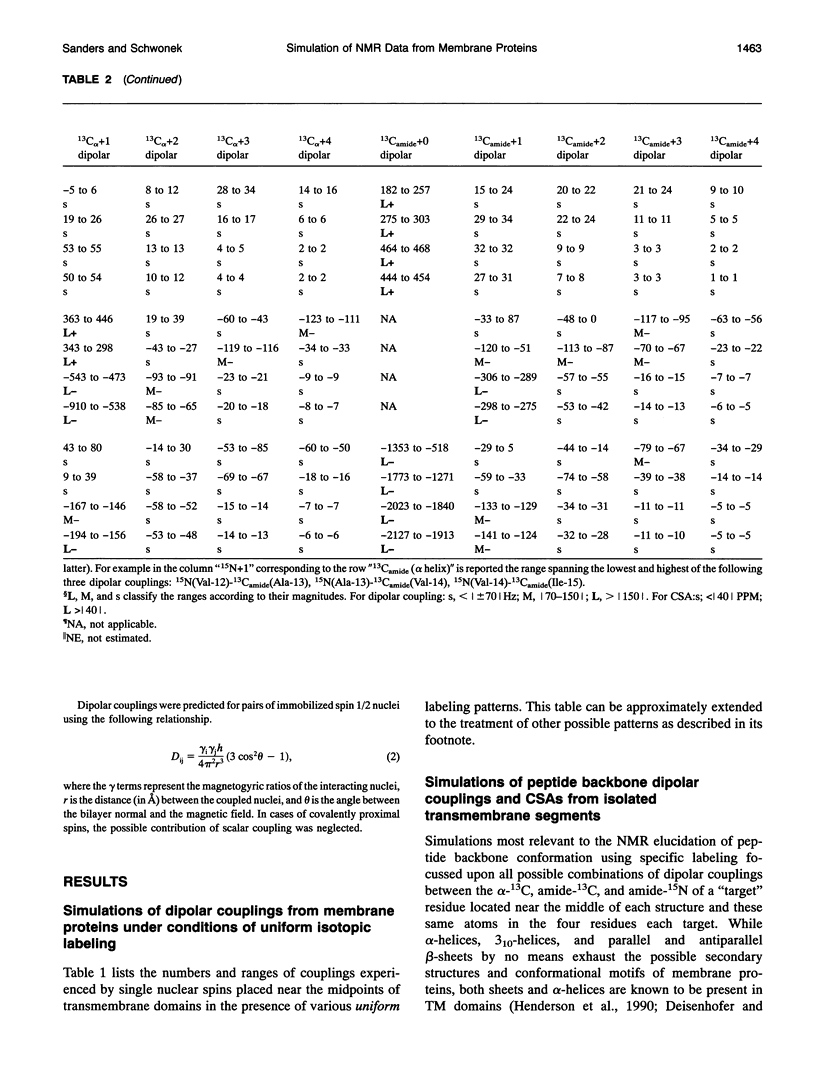
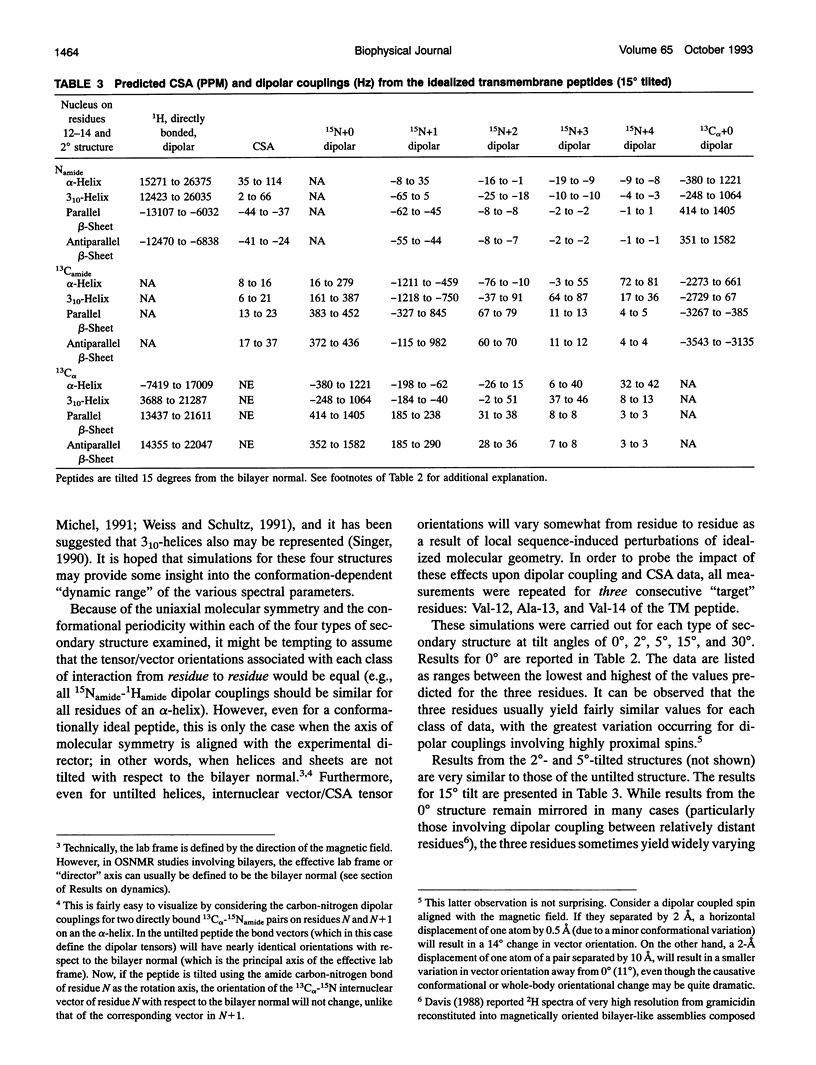
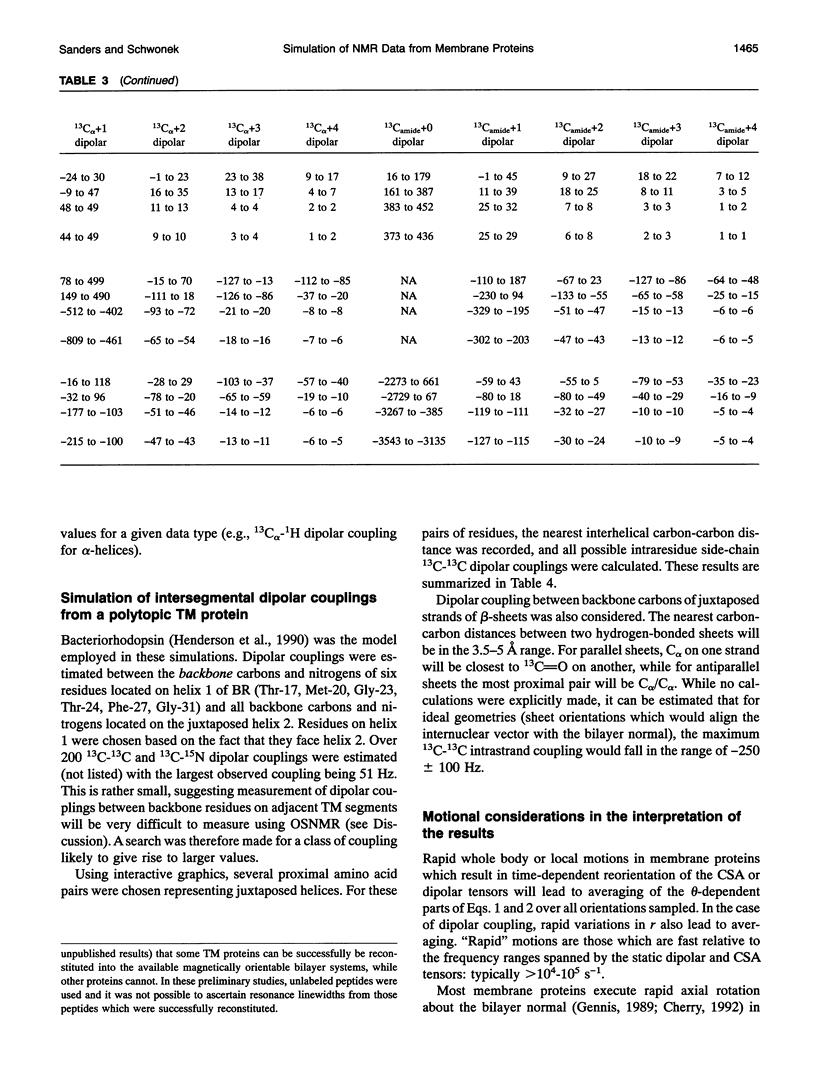
Several hundred solid state NMR dipolar couplings and chemical shift anisotropies were simulated for the polytopic membrane protein, bacteriorhodopsin, and for an idealized transmembrane peptide conforming to several different secondary structures (alpha- and 3(10)-helices and parallel and antiparallel beta-sheets), each at several tilt angles with respect to the bilayer normal. The use of macroscopically oriented samples was assumed. The results of these simulations suggest: (i) Because of the r-3 dependence of dipolar coupling, it is likely to prove difficult to successfully execute uniform isotopic enrichment strategies to generate large numbers of quantitatively interpretable structural measurements in oriented sample NMR studies of membrane proteins. (ii) There are a number of readily implementable specific isotopic labeling schemes which can yield data patterns sufficient to identify local secondary structure for transmembrane segments of idealized proteins which are tilted by < 10 degrees with respect to the bilayer normal. (iii) The measurement of dipolar coupling constants between 13C-, 19F-, and/or 3H-labeled side chains of proximal residues may prove effective as routes to long range tertiary structural data constraints.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chiu S. W., Nicholson L. K., Brenneman M. T., Subramaniam S., Teng Q., McCammon J. A., Cross T. A., Jakobsson E. Molecular dynamics computations and solid state nuclear magnetic resonance of the gramicidin cation channel. Biophys J. 1991 Oct;60(4):974–978. doi: 10.1016/S0006-3495(91)82131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell B. A., Separovic F., Baldassi A. J., Smith R. Conformation and orientation of gramicidin a in oriented phospholipid bilayers measured by solid state carbon-13 NMR. Biophys J. 1988 Jan;53(1):67–76. doi: 10.1016/S0006-3495(88)83066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. H. 2H nuclear magnetic resonance of exchange-labeled gramicidin in an oriented lyotropic nematic phase. Biochemistry. 1988 Jan 12;27(1):428–436. doi: 10.1021/bi00401a064. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. High-resolution structures of photosynthetic reaction centers. Annu Rev Biophys Biophys Chem. 1991;20:247–266. doi: 10.1146/annurev.bb.20.060191.001335. [DOI] [PubMed] [Google Scholar]
- Dufourc E. J., Mayer C., Stohrer J., Althoff G., Kothe G. Dynamics of phosphate head groups in biomembranes. Comprehensive analysis using phosphorus-31 nuclear magnetic resonance lineshape and relaxation time measurements. Biophys J. 1992 Jan;61(1):42–57. doi: 10.1016/S0006-3495(92)81814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare B. J., Howard K. P., Prestegard J. H. Torsion angle analysis of glycolipid order at membrane surfaces. Biophys J. 1993 Feb;64(2):392–398. doi: 10.1016/S0006-3495(93)81380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminga M. A., Sanders J. C., Spruijt R. B. Spectroscopy of lipid-protein interactions: structural aspects of two different forms of the coat protein of bacteriophage M13 incorporated in model membranes. Prog Lipid Res. 1992;31(3):301–333. doi: 10.1016/0163-7827(92)90011-7. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Herzfeld J., Mulliken C. M., Siminovitch D. J., Griffin R. G. Contrasting molecular dynamics in red and purple membrane fractions of the Halobacterium halobium. Biophys J. 1987 Nov;52(5):855–858. doi: 10.1016/S0006-3495(87)83278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian J. A., Taylor M. J., Koeppe R. E., 2nd Orientation of the valine-1 side chain of the gramicidin transmembrane channel and implications for channel functioning. A 2H NMR study. Biochemistry. 1992 Nov 24;31(46):11283–11290. doi: 10.1021/bi00161a004. [DOI] [PubMed] [Google Scholar]
- Leo G. C., Colnago L. A., Valentine K. G., Opella S. J. Dynamics of fd coat protein in lipid bilayers. Biochemistry. 1987 Feb 10;26(3):854–862. doi: 10.1021/bi00377a029. [DOI] [PubMed] [Google Scholar]
- Lewis B. A., Harbison G. S., Herzfeld J., Griffin R. G. NMR structural analysis of a membrane protein: bacteriorhodopsin peptide backbone orientation and motion. Biochemistry. 1985 Aug 13;24(17):4671–4679. doi: 10.1021/bi00338a029. [DOI] [PubMed] [Google Scholar]
- Li S. C., Deber C. M. Influence of glycine residues on peptide conformation in membrane environments. Int J Pept Protein Res. 1992 Sep-Oct;40(3-4):243–248. doi: 10.1111/j.1399-3011.1992.tb00297.x. [DOI] [PubMed] [Google Scholar]
- Loomis C. R., Walsh J. P., Bell R. M. sn-1,2-Diacylglycerol kinase of Escherichia coli. Purification, reconstitution, and partial amino- and carboxyl-terminal analysis. J Biol Chem. 1985 Apr 10;260(7):4091–4097. [PubMed] [Google Scholar]
- Mai W., Hu W., Wang C., Cross T. A. Orientational constraints as three-dimensional structural constraints from chemical shift anisotropy: the polypeptide backbone of gramicidin A in a lipid bilayer. Protein Sci. 1993 Apr;2(4):532–542. doi: 10.1002/pro.5560020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll F., 3rd, Cross T. A. Optimizing and characterizing alignment of oriented lipid bilayers containing gramicidin D. Biophys J. 1990 Feb;57(2):351–362. doi: 10.1016/S0006-3495(90)82536-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L. K., Teng Q., Cross T. A. Solid-state nuclear magnetic resonance derived model for dynamics in the polypeptide backbone of the gramicidin A channel. J Mol Biol. 1991 Apr 5;218(3):621–637. doi: 10.1016/0022-2836(91)90706-c. [DOI] [PubMed] [Google Scholar]
- Opella S. J., Stewart P. L. Solid-state nuclear magnetic resonance structural studies of proteins. Methods Enzymol. 1989;176:242–275. doi: 10.1016/0076-6879(89)76015-8. [DOI] [PubMed] [Google Scholar]
- Sami M., Dempsey C. Hydrogen exchange from the transbilayer hydrophobic peptide of glycophorin reconstituted in lipid bilayers. FEBS Lett. 1988 Nov 21;240(1-2):211–215. doi: 10.1016/0014-5793(88)80370-3. [DOI] [PubMed] [Google Scholar]
- Sanders C. R., 2nd, Schaff J. E., Prestegard J. H. Orientational behavior of phosphatidylcholine bilayers in the presence of aromatic amphiphiles and a magnetic field. Biophys J. 1993 Apr;64(4):1069–1080. doi: 10.1016/S0006-3495(93)81473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. R., 2nd, Schwonek J. P. An approximate model and empirical energy function for solute interactions with a water-phosphatidylcholine interface. Biophys J. 1993 Sep;65(3):1207–1218. doi: 10.1016/S0006-3495(93)81158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C. R., 2nd, Schwonek J. P. Characterization of magnetically orientable bilayers in mixtures of dihexanoylphosphatidylcholine and dimyristoylphosphatidylcholine by solid-state NMR. Biochemistry. 1992 Sep 22;31(37):8898–8905. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
- Sanders C. R., 2nd Solid state 13C NMR of unlabeled phosphatidylcholine bilayers: spectral assignments and measurement of carbon-phosphorus dipolar couplings and 13C chemical shift anisotropies. Biophys J. 1993 Jan;64(1):171–181. doi: 10.1016/S0006-3495(93)81352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon K. J., Kim Y., Colnago L. A., Opella S. J. NMR studies of the structure and dynamics of membrane-bound bacteriophage Pf1 coat protein. Science. 1991 May 31;252(5010):1303–1305. doi: 10.1126/science.1925542. [DOI] [PubMed] [Google Scholar]
- Singer S. J. The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- Smith R. L., Oldfield E. Dynamic structure of membranes by deuterium NMR. Science. 1984 Jul 20;225(4659):280–288. doi: 10.1126/science.6740310. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Peersen O. B. Solid-state NMR approaches for studying membrane protein structure. Annu Rev Biophys Biomol Struct. 1992;21:25–47. doi: 10.1146/annurev.bb.21.060192.000325. [DOI] [PubMed] [Google Scholar]
- Teng Q., Nicholson L. K., Cross T. A. Experimental determination of torsion angles in the polypeptide backbone of the gramicidin A channel by solid state nuclear magnetic resonance. J Mol Biol. 1991 Apr 5;218(3):607–619. doi: 10.1016/0022-2836(91)90705-b. [DOI] [PubMed] [Google Scholar]
- Weiss M. S., Schulz G. E. Structure of porin refined at 1.8 A resolution. J Mol Biol. 1992 Sep 20;227(2):493–509. doi: 10.1016/0022-2836(92)90903-w. [DOI] [PubMed] [Google Scholar]
- Williams K. A., Deber C. M. Proline residues in transmembrane helices: structural or dynamic role? Biochemistry. 1991 Sep 17;30(37):8919–8923. doi: 10.1021/bi00101a001. [DOI] [PubMed] [Google Scholar]


