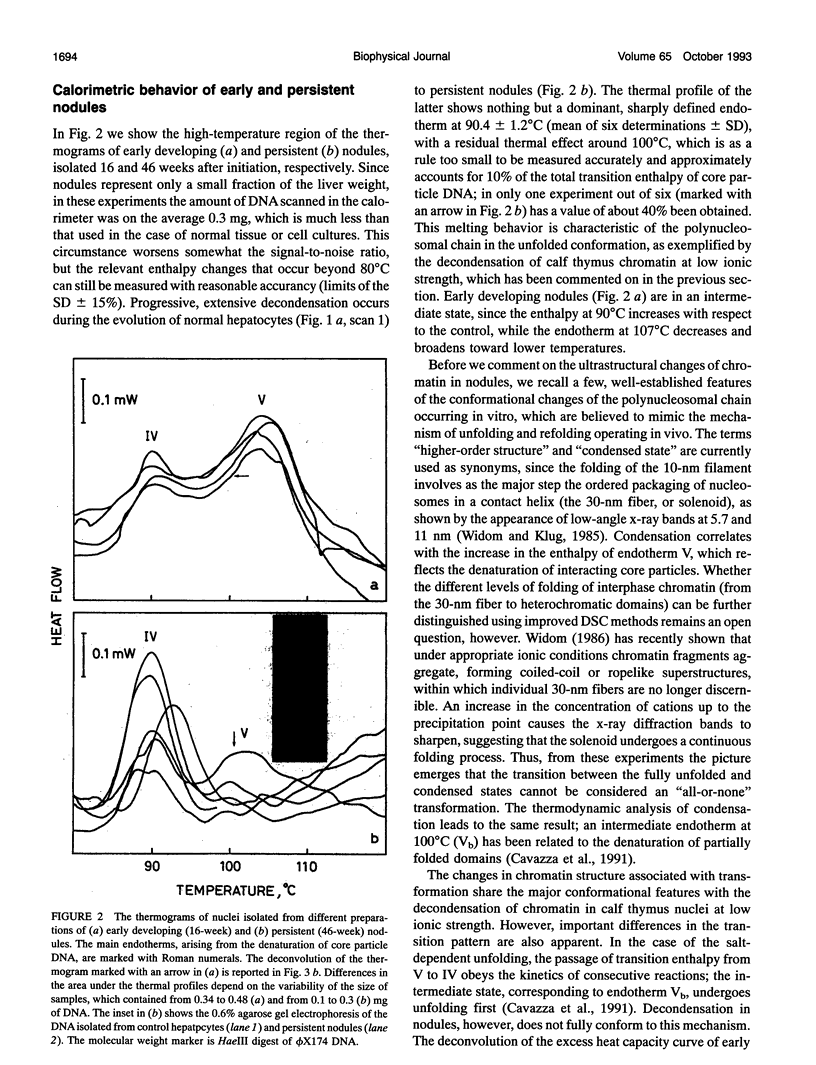
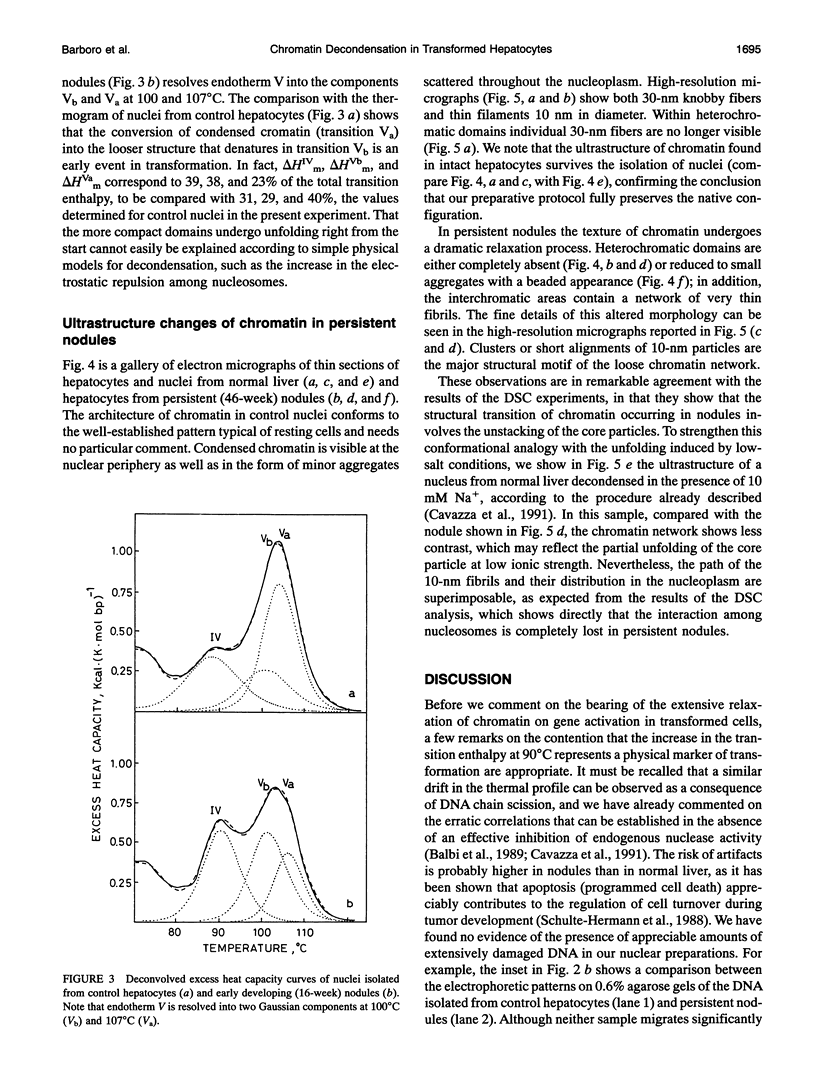
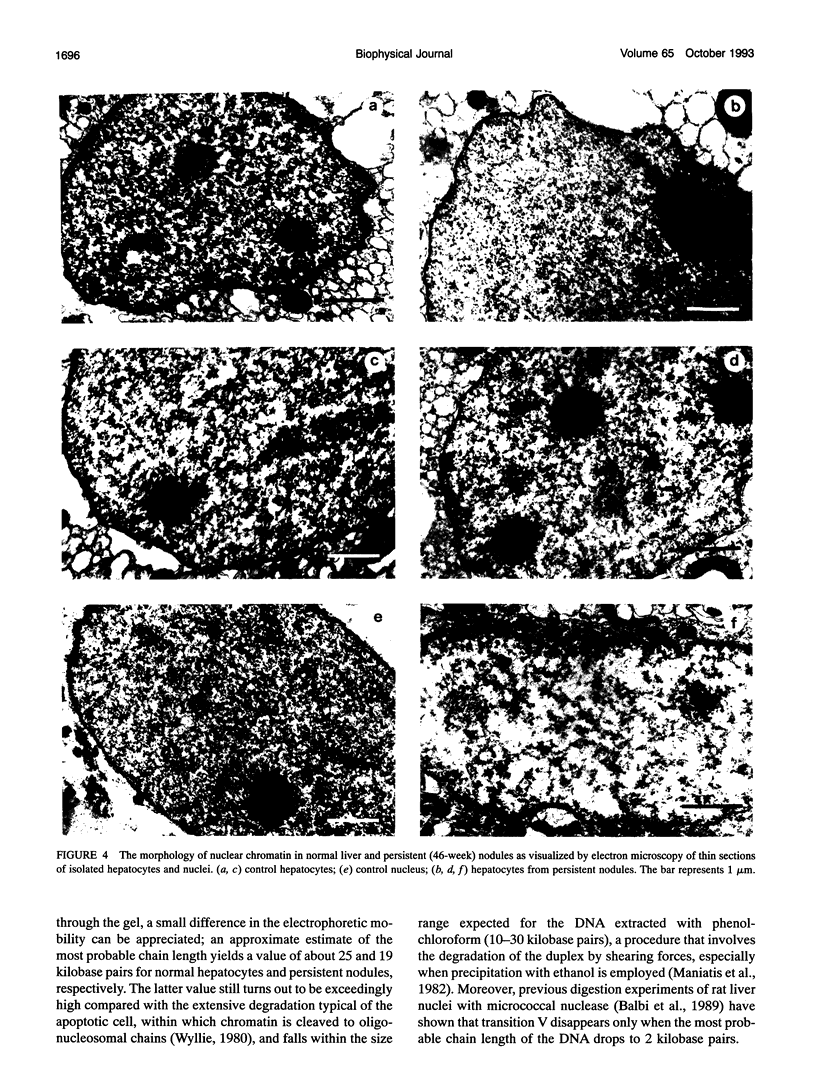
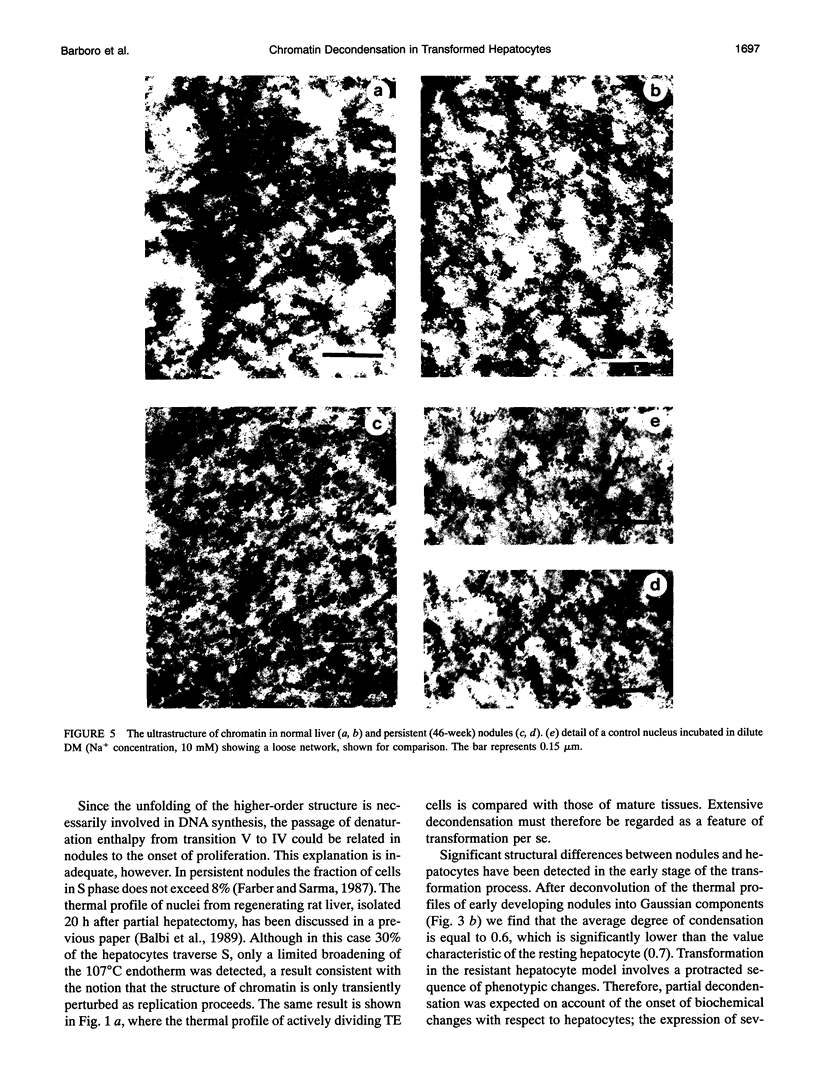
Abstract
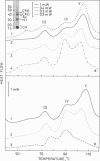
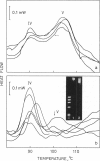
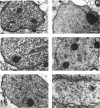
Using differential scanning calorimetry and complementary ultrastructural observations, we have characterized the status of chromatin during the transformation of rat hepatocytes in the resistant hepatocyte model of Solt and Farber (1976. Nature (Lond.). 263:701-703). Differential scanning calorimetry affords a measure of the degree of condensation of chromatin in situ and has therefore been used in this work for the purpose of establishing the nature of the structural changes associated with the emergence of successive cellular populations. Since the resistant hepatocyte model generates a series of synchronous phenotypic changes, it was possible to determine unambiguously the content of heterochromatin at each step of the process. The higher-order structure undergoes a partial relaxation in early developing nodules, isolated 16 weeks after initiation; the thermal transition at 90 degrees C, which is characteristic of noninteracting core particles, increases with respect to control hepatocytes. Dramatic changes occur in persistent (46-week) nodules. The 90 degrees C endotherm dominates the thermogram, while the transition at 107 degrees C, corresponding to the denaturation of the core particle packaged within the heterochromatic domains, disappears. The complete loss of the higher-order structure at this stage of transformation has been further verified by ultrastructural observations on thin nuclear sections. Ten-nm filaments, having a beaded appearance, are scattered throughout the nucleoplasm and clearly result from the decondensation of 30-nm-thick fibers. This catastrophic relaxation process cannot be related to an effective increase in gene activity. Rather, our observations suggest that during transformation chromatin is in a state of high transcriptional competence associated with the alert of general cellular programs. This view is consistent with the finding that in persistent nodules the DNA is extensively hypomethylated with respect to normal liver.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almagor M., Cole R. D. Changes in chromatin structure during the aging of cell cultures as revealed by differential scanning calorimetry. Biochemistry. 1989 Jun 27;28(13):5688–5693. doi: 10.1021/bi00439a052. [DOI] [PubMed] [Google Scholar]
- Balbi C., Abelmoschi M. L., Gogioso L., Parodi S., Barboro P., Cavazza B., Patrone E. Structural domains and conformational changes in nuclear chromatin: a quantitative thermodynamic approach by differential scanning calorimetry. Biochemistry. 1989 Apr 18;28(8):3220–3227. doi: 10.1021/bi00434a016. [DOI] [PubMed] [Google Scholar]
- Balbi C., Abelmoschi M. L., Zunino A., Cuniberti C., Cavazza B., Barboro P., Patrone E. The decondensation process of nuclear chromatin as investigated by differential scanning calorimetry. Biochem Pharmacol. 1988 May 1;37(9):1815–1816. doi: 10.1016/0006-2952(88)90460-1. [DOI] [PubMed] [Google Scholar]
- Cavazza B., Brizzolara G., Lazzarini G., Patrone E., Piccardo M., Barboro P., Parodi S., Pasini A., Balbi C. Thermodynamics of condensation of nuclear chromatin. A differential scanning calorimetry study of the salt-dependent structural transitions. Biochemistry. 1991 Sep 17;30(37):9060–9072. doi: 10.1021/bi00101a022. [DOI] [PubMed] [Google Scholar]
- Farber E., Sarma D. S. Hepatocarcinogenesis: a dynamic cellular perspective. Lab Invest. 1987 Jan;56(1):4–22. [PubMed] [Google Scholar]
- Franke W. W., Denk H., Kalt R., Schmid E. Biochemical and immunological identification of cytokeratin proteins present in hepatocytes of mammalian liver tissue. Exp Cell Res. 1981 Feb;131(2):299–318. doi: 10.1016/0014-4827(81)90234-2. [DOI] [PubMed] [Google Scholar]
- Fulmer A. W., Fasman G. D. Ionic strength-dependent conformational transitions of chromatin. Circular dichroism and thermal denaturation studies. Biopolymers. 1979 Nov;18(11):2875–2891. doi: 10.1002/bip.1979.360181115. [DOI] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Activation of cellular genes by avian RNA tumor viruses. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5351–5354. doi: 10.1073/pnas.77.9.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen J., Sistonen L., Alitalo K., Hölttä E. c-Ha-rasVal 12 oncogene-transformed NIH-3T3 fibroblasts display more decondensed nucleosomal organization than normal fibroblasts. J Cell Biol. 1990 Jul;111(1):9–17. doi: 10.1083/jcb.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardson K. E., Levy S. B. Chromatin reorganization during emergence of malignant Friend tumors: early changes in H2A and H2B variants and nucleosome repeat length. Exp Cell Res. 1989 Jan;180(1):209–219. doi: 10.1016/0014-4827(89)90225-5. [DOI] [PubMed] [Google Scholar]
- Nicolini C., Trefiletti V., Cavazza B., Cuniberti C., Patrone E., Carlo P., Brambilla G. Quaternary and quinternary structures of native chromatin DNA in liver nuclei: differential scanning calorimetry. Science. 1983 Jan 14;219(4581):176–178. doi: 10.1126/science.6849127. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Stereo electron microscopy of the 25-nm chromatin fibers in isolated nuclei. J Cell Biol. 1979 Apr;81(1):260–265. doi: 10.1083/jcb.81.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi S., Pala M., Russo P., Balbi C., Abelmoschi M. L., Taningher M., Zunino A., Ottaggio L., de Ferrari M., Carbone A. Alkaline DNA fragmentation, DNA disentanglement evaluated viscosimetrically and sister chromatid exchanges, after treatment in vivo with nitrofurantoin. Chem Biol Interact. 1983 Jul 1;45(1):77–94. doi: 10.1016/0009-2797(83)90044-3. [DOI] [PubMed] [Google Scholar]
- Reuter G., Giarre M., Farah J., Gausz J., Spierer A., Spierer P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990 Mar 15;344(6263):219–223. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- Semple-Roberts E., Hayes M. A., Armstrong D., Becker R. A., Racz W. J., Farber E. Alternative methods of selecting rat hepatocellular nodules resistant to 2-acetylaminofluorene. Int J Cancer. 1987 Nov 15;40(5):643–645. doi: 10.1002/ijc.2910400512. [DOI] [PubMed] [Google Scholar]
- Staynov D. Z. Thermal denaturation profiles and the structure of chromatin. Nature. 1976 Dec 9;264(5586):522–525. doi: 10.1038/264522a0. [DOI] [PubMed] [Google Scholar]
- Svaren J., Chalkley R. The structure and assembly of active chromatin. Trends Genet. 1990 Feb;6(2):52–56. doi: 10.1016/0168-9525(90)90074-g. [DOI] [PubMed] [Google Scholar]
- Touchette N. A., Anton E., Cole R. D. A higher order chromatin structure that is lost during differentiation of mouse neuroblastoma cells. J Biol Chem. 1986 Feb 15;261(5):2185–2188. [PubMed] [Google Scholar]
- Touchette N. A., Cole R. D. Differential scanning calorimetry of nuclei reveals the loss of major structural features in chromatin by brief nuclease treatment. Proc Natl Acad Sci U S A. 1985 May;82(9):2642–2646. doi: 10.1073/pnas.82.9.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tökés Z. A., Clawson G. A. Proteolytic activity associated with the nuclear scaffold. The effect of self-digestion on lamins. J Biol Chem. 1989 Sep 5;264(25):15059–15065. [PubMed] [Google Scholar]
- Villeponteau B., Brawley J., Martinson H. G. Nucleosome spacing is compressed in active chromatin domains of chick erythroid cells. Biochemistry. 1992 Feb 11;31(5):1554–1563. doi: 10.1021/bi00120a037. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Tissue-specific gene expression and chromatin structure. Harvey Lect. 1983 1984;79:217–244. [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Tatchell K., Van Holde K. E., Klump H. Thermal denaturation of nucleosomal core particles. Nucleic Acids Res. 1978 Jan;5(1):139–160. doi: 10.1093/nar/5.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J., Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985 Nov;43(1):207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- Widom J. Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. J Mol Biol. 1986 Aug 5;190(3):411–424. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]