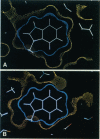
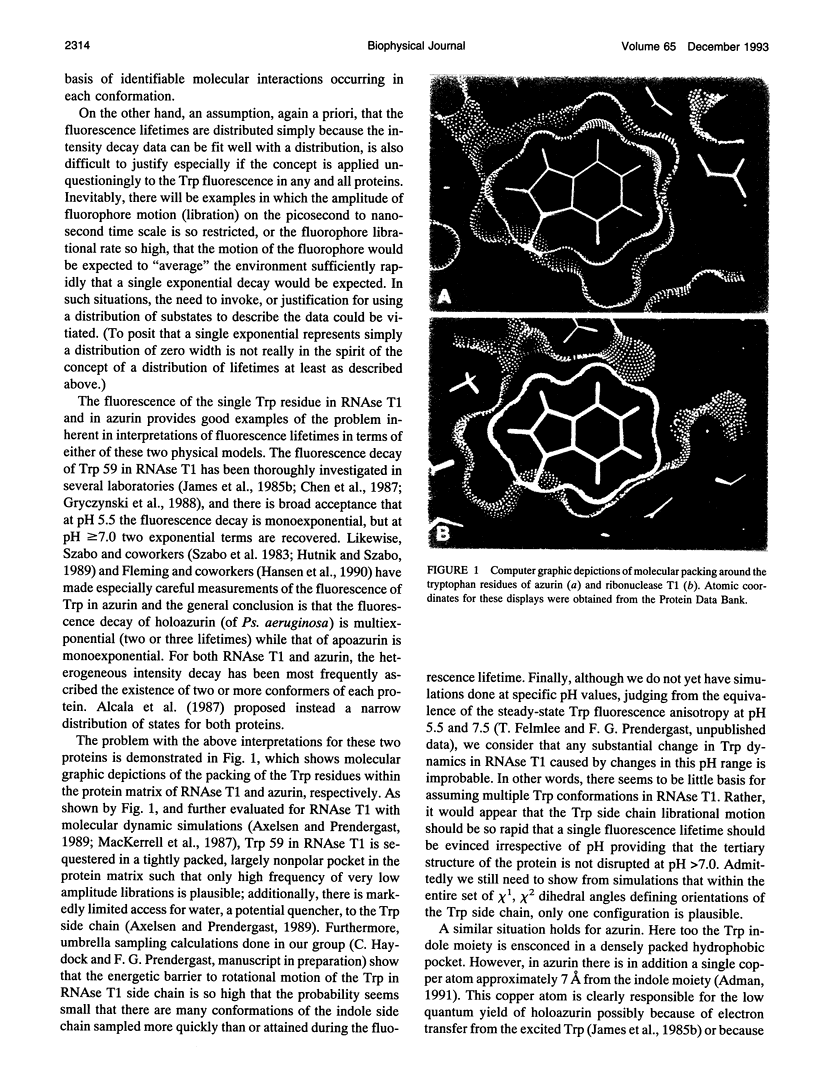
Abstract
Tryptophan fluorescence intensity decay in proteins is modeled by multiexponential functions characterized by lifetimes and preexponential factors. Commonly, multiple conformations of the protein are invoked to explain the recovery of two or more lifetimes from the experimental data. However, in many proteins the structure seems to preclude the possibility of multiple conformers sufficiently different from one another to justify such an inference. We present here another plausible multiexponential model based on the assumption that an energetically excited donor surrounded by N acceptor molecules decays by specific radiative and radiationless relaxation processes, and by transferring its energy to acceptors present in or close to the protein matrix. If interactions between the acceptors themselves and back energy transfer are neglected, we show that the intensity decay function contain 2N exponential components characterized by the unperturbed donor lifetime, by energy transfer rates and a probability of occurrence for the corresponding process. We applied this model to the fluorescence decay of holo- and apoazurin, ribonuclease T1, and the reduced single tryptophan mutant (W28F) of thioredoxin. Use of a multiexponential model for the analysis of the fluorescence intensity decay can therefore be justified, without invoking multiple protein conformations.
Full text
PDF

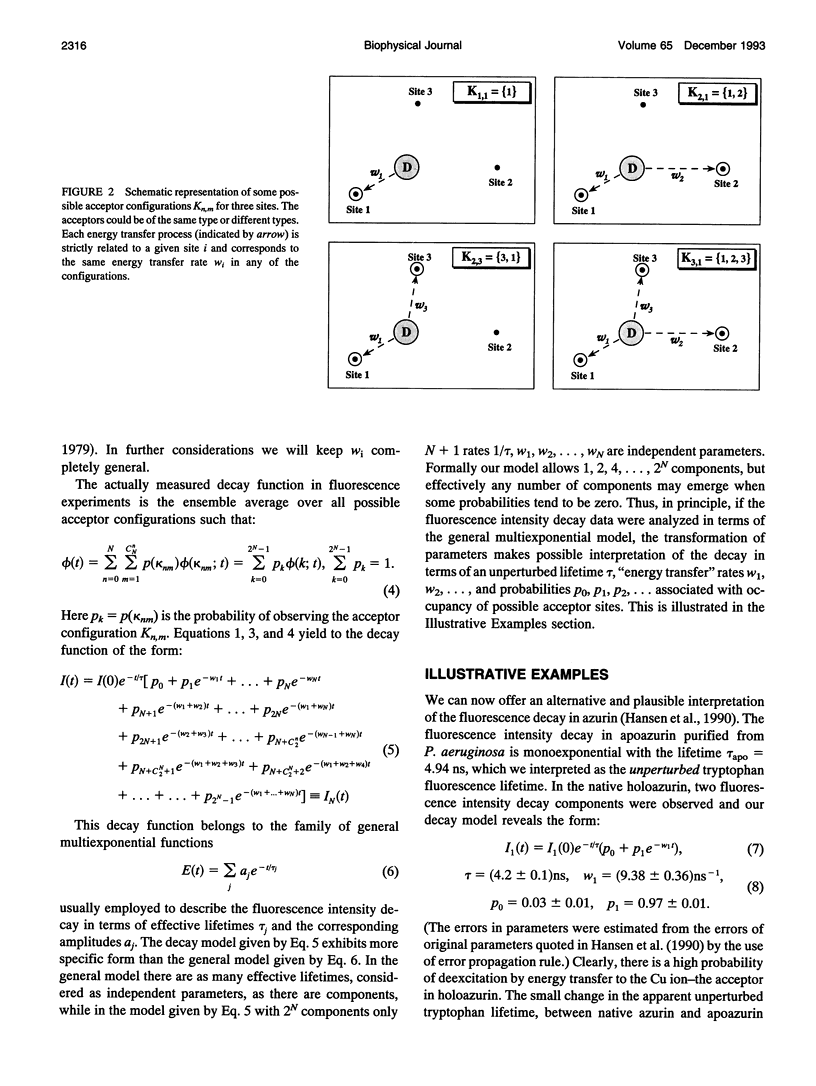

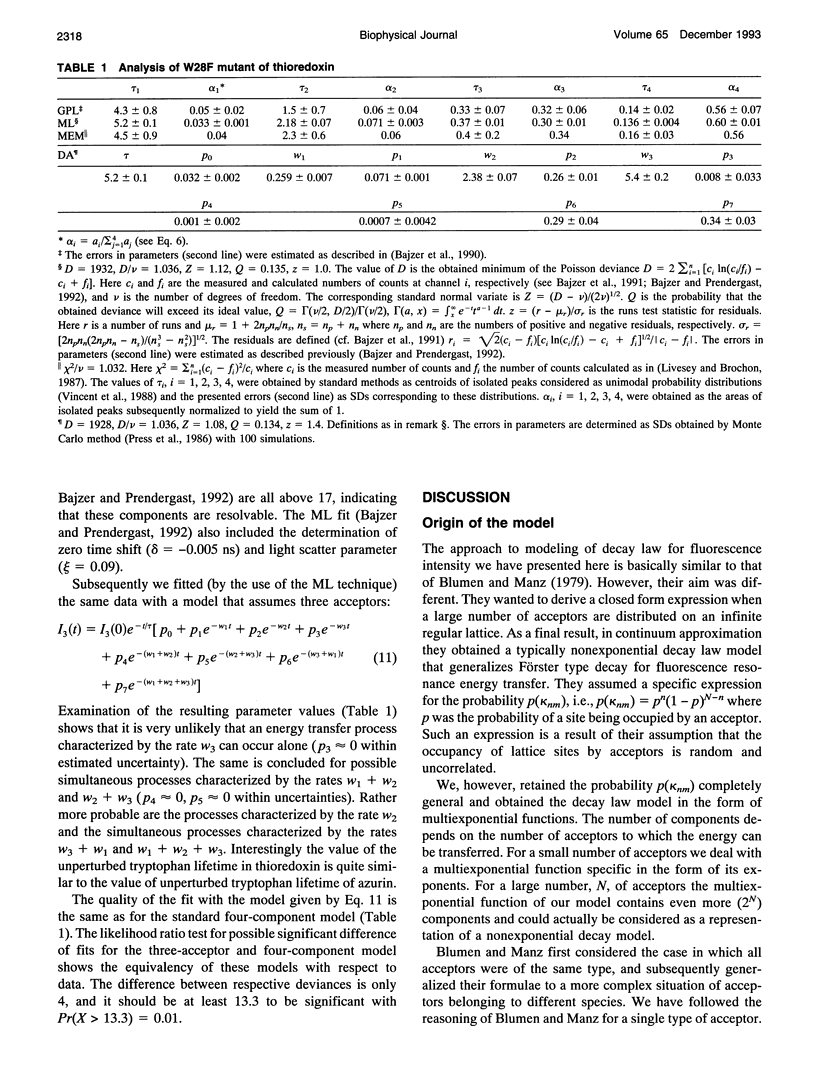




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T. Copper protein structures. Adv Protein Chem. 1991;42:145–197. doi: 10.1016/s0065-3233(08)60536-7. [DOI] [PubMed] [Google Scholar]
- Alcala J. R., Gratton E., Prendergast F. G. Interpretation of fluorescence decays in proteins using continuous lifetime distributions. Biophys J. 1987 Jun;51(6):925–936. doi: 10.1016/S0006-3495(87)83420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen P. H., Prendergast F. G. Molecular dynamics of tryptophan in ribonuclease-T1. II. Correlations with fluorescence. Biophys J. 1989 Jul;56(1):43–66. doi: 10.1016/S0006-3495(89)82651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajzer Z., Prendergast F. G. Maximum likelihood analysis of fluorescence data. Methods Enzymol. 1992;210:200–237. doi: 10.1016/0076-6879(92)10012-3. [DOI] [PubMed] [Google Scholar]
- Bajzer Z., Sharp J. C., Sedarous S. S., Prendergast F. G. Padé-Laplace method for the analysis of time-resolved fluorescence decay curves. Eur Biophys J. 1990;18(2):101–115. doi: 10.1007/BF00183269. [DOI] [PubMed] [Google Scholar]
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Chen L. X., Longworth J. W., Fleming G. R. Picosecond time-resolved fluorescence of ribonuclease T1. A pH and substrate analogue binding study. Biophys J. 1987 Jun;51(6):865–873. doi: 10.1016/S0006-3495(87)83414-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. F., Knutson J. R., Ziffer H., Porter D. Fluorescence of tryptophan dipeptides: correlations with the rotamer model. Biochemistry. 1991 May 28;30(21):5184–5195. doi: 10.1021/bi00235a011. [DOI] [PubMed] [Google Scholar]
- Elofsson A., Rigler R., Nilsson L., Roslund J., Krause G., Holmgren A. Motion of aromatic side chains, picosecond fluorescence, and internal energy transfer in Escherichia coli thioredoxin studied by site-directed mutagenesis, time-resolved fluorescence spectroscopy, and molecular dynamics simulations. Biochemistry. 1991 Oct 8;30(40):9648–9656. doi: 10.1021/bi00104a012. [DOI] [PubMed] [Google Scholar]
- Gryczynski I., Eftink M., Lakowicz J. R. Conformation heterogeneity in proteins as an origin of heterogeneous fluorescence decays, illustrated by native and denatured ribonuclease T1. Biochim Biophys Acta. 1988 Jun 13;954(3):244–252. doi: 10.1016/0167-4838(88)90079-9. [DOI] [PubMed] [Google Scholar]
- Hansen J. E., Longworth J. W., Fleming G. R. Photophysics of metalloazurins. Biochemistry. 1990 Aug 7;29(31):7329–7338. doi: 10.1021/bi00483a024. [DOI] [PubMed] [Google Scholar]
- Hochstrasser R. M., Negus D. K. Picosecond fluorescence decay of tryptophans in myoglobin. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4399–4403. doi: 10.1073/pnas.81.14.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Tryptophan fluorescence study of conformational transitions of the oxidized and reduced form of thioredoxin. J Biol Chem. 1972 Apr 10;247(7):1992–1998. [PubMed] [Google Scholar]
- Hutnik C. M., Szabo A. G. Confirmation that multiexponential fluorescence decay behavior of holoazurin originates from conformational heterogeneity. Biochemistry. 1989 May 2;28(9):3923–3934. doi: 10.1021/bi00435a045. [DOI] [PubMed] [Google Scholar]
- James D. R., Demmer D. R., Steer R. P., Verrall R. E. Fluorescence lifetime quenching and anisotropy studies of ribonuclease T1. Biochemistry. 1985 Sep 24;24(20):5517–5526. doi: 10.1021/bi00341a036. [DOI] [PubMed] [Google Scholar]
- Klafter J., Shlesinger M. F. On the relationship among three theories of relaxation in disordered systems. Proc Natl Acad Sci U S A. 1986 Feb;83(4):848–851. doi: 10.1073/pnas.83.4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey A. K., Brochon J. C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys J. 1987 Nov;52(5):693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A. D., Jr, Rigler R., Nilsson L., Hahn U., Saenger W. Protein dynamics. A time-resolved fluorescence, energetic and molecular dynamics study of ribonuclease T1. Biophys Chem. 1987 May 9;26(2-3):247–261. doi: 10.1016/0301-4622(87)80027-3. [DOI] [PubMed] [Google Scholar]
- Mérola F., Rigler R., Holmgren A., Brochon J. C. Picosecond tryptophan fluorescence of thioredoxin: evidence for discrete species in slow exchange. Biochemistry. 1989 Apr 18;28(8):3383–3398. doi: 10.1021/bi00434a038. [DOI] [PubMed] [Google Scholar]
- Petrich J. W., Longworth J. W., Fleming G. R. Internal motion and electron transfer in proteins: a picosecond fluorescence study of three homologous azurins. Biochemistry. 1987 May 19;26(10):2711–2722. doi: 10.1021/bi00384a010. [DOI] [PubMed] [Google Scholar]
- Ross J. B., Wyssbrod H. R., Porter R. A., Schwartz G. P., Michaels C. A., Laws W. R. Correlation of tryptophan fluorescence intensity decay parameters with 1H NMR-determined rotamer conformations: [tryptophan2]oxytocin. Biochemistry. 1992 Feb 18;31(6):1585–1594. doi: 10.1021/bi00121a002. [DOI] [PubMed] [Google Scholar]
- Szabo A. G., Stepanik T. M., Wayner D. M., Young N. M. Conformational heterogeneity of the copper binding site in azurin. A time-resolved fluorescence study. Biophys J. 1983 Mar;41(3):233–244. doi: 10.1016/S0006-3495(83)84433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Brochon J. C., Merola F., Jordi W., Gallay J. Nanosecond dynamics of horse heart apocytochrome c in aqueous solution as studied by time-resolved fluorescence of the single tryptophan residue (Trp-59). Biochemistry. 1988 Nov 29;27(24):8752–8761. doi: 10.1021/bi00424a010. [DOI] [PubMed] [Google Scholar]
- Weaver A. J., Kemple M. D., Prendergast F. G. Tryptophan sidechain dynamics in hydrophobic oligopeptides determined by use of 13C nuclear magnetic resonance spectroscopy. Biophys J. 1988 Jul;54(1):1–15. doi: 10.1016/S0006-3495(88)82925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]