Abstract
Endoplasmic reticulum (ER)p61, ERp72, and protein disulfide isomerase (PDI), which are members of the PDI family protein, are ubiquitously present in mammalian cells and are thought to participate in disulfide bond formation and isomerization. However, why the 3 different members need to be colocalized in the ER remains an enigma. We hypothesized that each PDI family protein might have different modes of enzymatic activity in disulfide bond formation and isomerization. We purified PDI, ERp61, and ERp72 proteins from rat liver microsomes and compared the effects of each protein on the folding of bovine pancreatic trypsin inhibitor (BPTI). ERp61 and ERp72 accelerated the initial steps more efficiently than did PDI. ERp61 and ERp72, however, accelerated the rate-limiting step less efficiently than did PDI. PDI or ERp72 did not impede the folding of BPTI by each other but rather catalyzed the folding reaction cooperatively with each other. These data suggest that differential enzymatic activities of ERp proteins and PDI represent a complementary contribution of these enzymes to protein folding in the ER.
INTRODUCTION
In the endoplasmic reticulum (ER), 2 classes of proteins assist polypeptide and protein folding (Gething and Sambrook 1992; Hammond and Helenius 1995; Trombetta and Parodi 2003): (1) a group of folding enzymes, such as peptidyl prolyl cis-trans isomerase and protein disulfide isomerase (PDI), the latter containing 2 thioredoxin homology units, which catalyze rate-limiting isomerization steps in protein folding (Gilbert 1998; Frand et al 2000; Freedman et al 2002; Sevier and Kaiser 2002) and (2) a group of molecular chaperones that stabilize unfolded or partially folded intermediates and prevent the formation of inappropriate folding interactions (Pfeffer and Rothman 1987; Johnson and van Waes 1999; Rapoport et al 1999). PDI and its related proteins, ERp61 and ERp72, comprise a PDI family (Lewis et al 1986; Mazzarella et al 1990; Martin et al 1991; Srivastava et al 1991, 1993; Urade et al 1992). ERp61 and ERp72 have been identified as ER proteins containing 2 and 3 thioredoxin homology units, respectively. The homology unit contains a Trp-Cys-Gly-His-Cys-Lys (WCGHCK) motif identical to the active site consensus sequences of PDI (Lee 1981; Mazzarella et al 1990, 1994; Gilbert 1998).
PDI catalyzes the formation and reduction of disulfide bonds and their isomerization in a wide range of substrate proteins in vitro, such as insulin, ribonuclease, bovine pancreatic trypsin inhibitor (BPTI), and in immunoglobulin Fab fragments (for a review see Creighton 1997 and Ruddon and Bedows 1997). PDI also catalyzes as a chaperone the renaturation of denatured, reduced insulin (Freedman et al 1989) or of ribonuclease T1 or AIII (Freedman 1989). The in vitro protein folding of BPTI is one of the best-characterized disulfide folding pathways. PDI increases by a factor of 3000–6000, the rates of kinetically trapped BPTI-folding intermediates (Weissman and Kim 1993).
The distributions of PDI, ERp61, and ERp72 have been compared in various tissues (Kozaki et al 1994; Marcus et al 1996). The tissue distributions of PDI and ERp72 messenger RNA (mRNAs) are similar, although ERp72 is not as abundant as is PDI in tissues, and the tissue distribution of ERp61 mRNA is markedly different from that of PDI and ERp72 mRNA (Marcus et al 1996). We previously studied the distribution of PDI family proteins in rat tissues and compared their localization by immunohistochemical double staining (Kozaki et al 1994; Iida et al 1996). Although these proteins were ubiquitously expressed in a wide variety of cell types, the extent of their expression varied in different cell types. Especially, a distinct difference in expression levels of the 3 proteins was observed between plasma cells, pancreatic islet cells, and goblet cells.
These ERp proteins can catalyze the reduction of the insulin disulfide bond (Füllekrug et al 1994; Kozaki et al 1994) or the reappearance of the biological activity of the denatured, reduced Fab fragment of immunoglobulin (Rupp et al 1994), but these activities also seem to be several fold lower than the activity of PDI (Füllekrug et al 1994; Rupp et al 1994). We previously estimated the activities of ERp61 and ERp72 as one-fifth the activity of PDI, using insulin reduction assay (Kozaki et al 1994).
Although ERp61 and ERp72 have a relatively lower enzymatic activity, they exist in the ER together with PDI. A good explanation does not exist regarding why all 3 PDI family proteins are colocalized in the ER. And why the expression level of ERp61 or ERp72 relative to that of PDI varies depending on cell or tissue type is not clear.
In this study, we hypothesized that each PDI family protein might have different modes of enzymatic activity in disulfide bond formation and isomerization and that they might have a cooperative interaction in the protein folding pathway. Using an in vitro BPTI-folding assay system, we studied how ERp61 and ERp72 function in a protein folding pathway.
MATERIALS AND METHODS
Reagents
BPTI (Trasylol™) was purchased from Bayer (Leverkusen, Germany), glutathione reduced (GSH) from Wako Pure Chemicals (Osaka, Japan), and glutathione disulfide (GSSG) from Sigma (St Louis, MO, USA). Diethylaminoethyl(DEAE)-sephacel, chelating-sepharose, heparin-sepharose, and monoQ column were purchased from Pharmacia (Uppsala, Sweden). The other reagents were purchased from Nakalai Tesque (Kyoto, Japan), Wako Pure Chemicals, and Katayama Chemicals (Osaka, Japan).
Preparation of PDI, ERp61, and ERp72
PDI, ERp61, and ERp72 were prepared from rat liver microsomes as described previously (Kozaki et al 1994) but with a minor modification. Microsomes were prepared as described previously (Walter and Blobel 1983; Kassenbrock and Kelly 1989). The microsomes were solubilized in 1% Triton X-100 and 0.5% sodium deoxycholate, ultracentrifuged, and the supernatants collected. The extracts were applied to DEAE-sephacel and were eluted with the gradient from 0 to 0.4 M NaCl. ERp proteins were eluted in the following order: ERp61, then PDI, and finally ERp72. The fractions containing PDI, ERp61, and ERp72 were confirmed by immunoblotting using specific antibodies against each ERp protein (Kozaki et al 1994). Fractions containing each ERp protein were separately applied to a monoQ column to concentrate the ER proteins. Fractions containing ERp61 or ERp72 were then applied to heparin-sepharose and were eluted with high ionic buffer. Fractions containing PDI were applied to a metal-chelating column (Zn2+-chelated) and were eluted with a high ionic buffer (Baksh et al 1995). The buffers of fractions containing each ERp protein were concentrated and exchanged using a centricon concentrator. The purity of each protein was confirmed by detecting as a single band on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Fig 1A).
Fig 1.
Preparation of PDI family proteins and the pathway of BPTI folding. (A) PDI, ERp61, and ERp72 were prepared from rat liver microsomes as described in Materials and Methods. The proteins were observed by coomassie brilliant blue R-250 staining. Lane 1: PDI; lane 2: ERp61; lane 3: ERp72
Purification and folding assay of BPTI
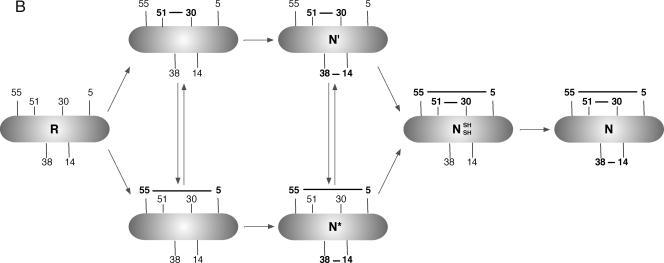
BPTI was purified in the reduced form on a Vydac C-18 preparative column, as described previously (Weissman and Kim 1991, 1992, 1993). The folding reactions of reduced BPTI in the presence or absence of ERp protein(s) were monitored (Weissman and Kim 1991). In brief (Baksh et al 1995), folding was initiated by adding redox buffer of degassed folding buffer (150 mM NaCl, 100 mM sodium phosphate, 1 mM ethylene diamine tetraacetate, pH 7.3, 2.0 mM GSH, 0.5 mM GSSG) either with or without 1.5 μM PDI family protein to reduced BPTI (the concentration of reduced BPTI in the reaction was finally 30 μM) (Bourdi et al 1995); at the indicated times, a portion of the folding reaction was quenched with 0.1 volume of 3 M HCl (Creighton 1997); the spectrum of intermediates present at each time was determined by high-performance liquid chromatography (HPLC) in an acidic condition (pH 2). The disulfide linkages of each intermediate were determined according to the data reported by Weissman and Kim (1991, 1993), and folding pathway of BPTI is represented in Figure 1B. All folding experiments were performed in a thermal incubator at 25°C. The redox buffer was chosen to mimic the redox conditions of the ER.
Fig 1.
(Continued) (B) Schematic representation of the folding pathway for BPTI. PDI, protein disulfide isomerase; BPTI, bovine pancreatic trypsin inhibitor; ER, endoplasmic reticulum
Acid quenching, condition for separation, and folding assay of intermediates
Acid quenching was achieved by adding of 0.1 volume of 3 M HCl to give a final pH of approximately 2. Acid-quenched intermediates were purified by HPLC and lyophilized as described in Weissman and Kim (1991). When a delay between acid quenching and HPLC analysis was necessary, the experiment was repeated with the order of chromatography prepared to show that a rearrangement did not occur while the mix awaited separation. The gradient used for the HPLC separation was 0 minutes, 90% A; 15 minutes, 75% A; 35 minutes, 73% A; 50 minutes, 72% A; and 140 minutes, 69% A. The folding reactions of intermediates, N′ and N*, in the presence or absence of the PDI family protein were monitored as described in the previous section. The concentrations of intermediates and the PDI family protein in the reaction were finally 30 and 1.5 μM, respectively. The concentrations of PDI and ERp72 for performance of cooperative folding of BPTI varied from 0.375 to 1.125 μM but were adjusted so that the total amount of PDI and ERp72 in the reaction was equal to that in the reaction with PDI or ERp72 alone. Changes in the amount of fully folded BPTI were statistically evaluated using the analysis of variance program of STATISTICA (StatSoft, Tulsa, OK, USA). Post hoc tests were performed using Tukey's honestly significant difference (HSD) test.
RESULTS
ERp61 and ERp72 accelerated the folding of native BPTI but less effectively than PDI
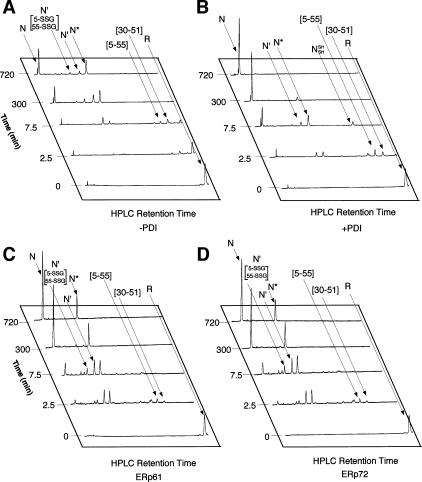
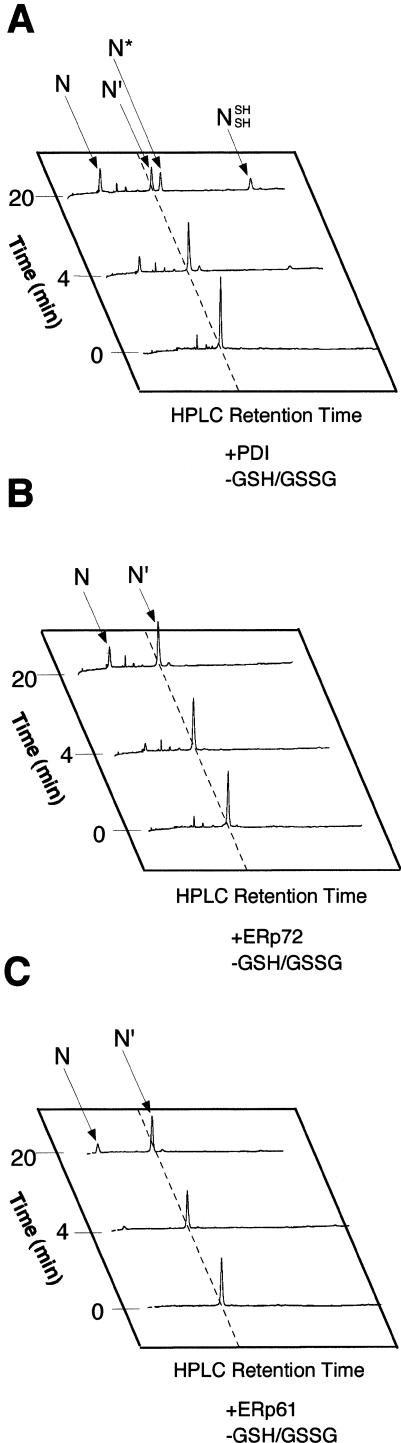
PDI dramatically increased both the yield and rate of formation of native BPTI in a physiological redox buffer (Fig 2). The result was similar to that reported previously (Weissman and Kim 1993). The rate of formation of the 2 kinetically trapped native intermediates N* and N′, which contained 2 native disulfide bonds [5–55; 14–38] and [30–51; 14–38], respectively, increased moderately (about 3-fold) by PDI (Fig 2B). The striking feature of the PDI-catalyzed folding reaction was that N′ and N* intermediates appeared to be converted readily to native BPTI (N).
Fig 2.
Time course of folding of reduced BPTI in the presence of ERp protein with both 2.0 mM GSH and 0.5 mM GSSG. N′ [5-SSG; 55-SSG] denotes the double-mixed disulfide derivative of N′, in which Cys 5 and Cys 55 are each disulfide bonded to glutathione. The reactions with no enzyme (A), with PDI (B), with ERp61 (C), and with ERp72 (D) are indicated. BPTI, bovine pancreatic trypsin inhibitor; GSSG, glutathione disulfide; PDI, protein disulfide isomerase; ER, endoplasmic reticulum
To analyze whether the other PDI family proteins, ERp61 and ERp72, increase the yield and rate of formation of native BPTI as efficiently as does PDI, fully reduced BPTI was incubated with ERp61 or ERp72 instead of PDI under the same conditions (Fig 2). ERp61 and ERp72 catalyzed the folding pathway from the reduced form to either N* or N′ (Fig 2 C,D). The further folding steps from the intermediate(s) to the native form were also catalyzed compared with nonenzymatic reactions (Fig 2 A,C,D). But N* accumulated and did not decrease in amount between 300 and 720 minutes after the initiation of the reaction, when most of the 2 intermediates were eliminated in the reaction in the presence of PDI.
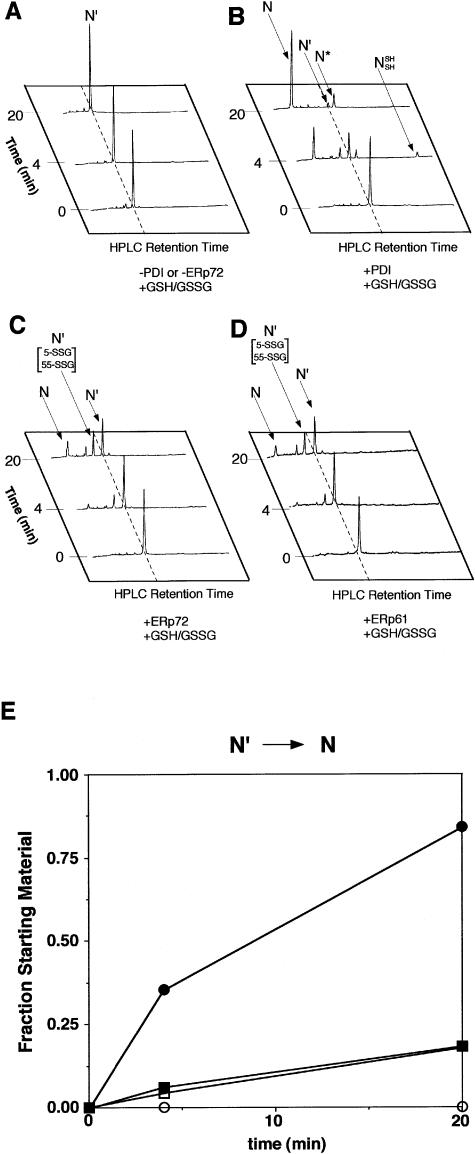
ERp61 and ERp72 efficiently catalyzed the rearrangement of the intermediate form N′ but not N*
To investigate the ability of ERp61 and ERp72 to fold 2 intermediate forms, N′ and N*, to native BPTI, we purified these intermediates and incubated each intermediate with PDI family protein. In the absence of PDI or ERp proteins, N′ did not fold to the native form during the incubation time (Fig 3A). PDI dramatically increased the rate of disulfide bond formation (Fig 3B). ERp72 moderately increased the rate of disulfide bond formation (Fig 3C). ERp61 had a similar effect on the folding of BPTI (Fig 3D). The folding of BPTI in the reaction catalyzed by ERp61 and ERp72 was almost to the same degree as that by PDI. NSHSH was observed at 4 minutes after the initiation of the reaction with PDI. In the reaction with ERp61 or ERp72, however, the intermediate [30–51; 14–38; 5-SSG; 55-SSG] was observed instead of NSHSH (Fig 3 C,D).
Fig 3.
Catalysis by ERp protein of the intramolecular rearrangement of the native 2-disulfide intermediate, N′, with both 2.0 mM GSH and 0.5 mM GSSG. The reaction with no enzyme (A), with PDI (B), with ERp72 (C), and with ERp61 (D) are indicated. (E) The kinetics of the native form:total BPTI ratio. The area under the curve of elution profiles corresponding to intermediates (N′, N*, and NSHSH) and native form (N) was measured using National Institutes of Health image. Molecular ratios are indicated in arbitrary values. ERp61 and ERp72 accelerated the conversion of N′ to N, but less efficiently than did PDI, and hardly converted N′ to N*. Open circles, no enzyme; closed circles, 1.5 μM PDI; open squares, 1.5 μM ERp61; closed squares, 1.5 μM ERp72. GSSG, glutathione disulfide; PDI, protein disulfide isomerase; ER, endoplasmic reticulum
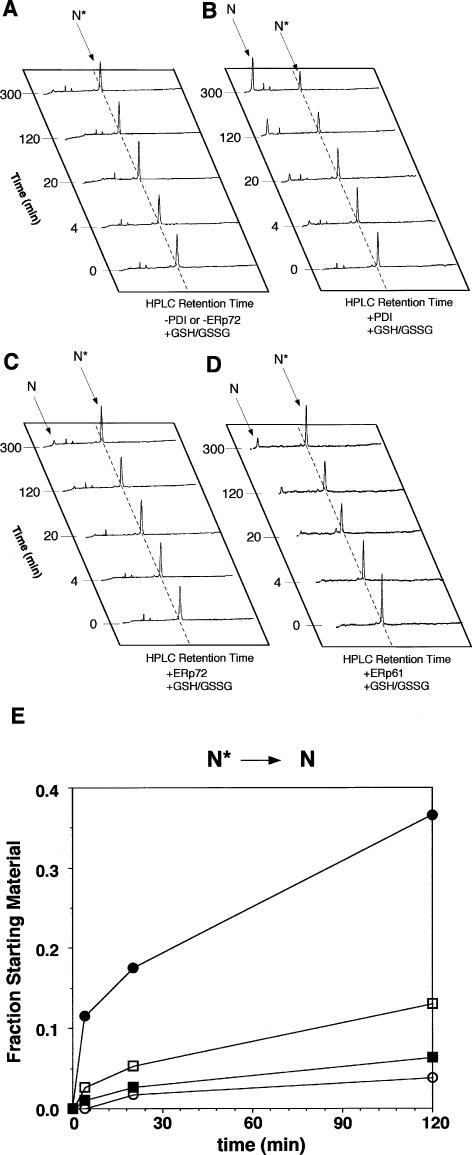
PDI increased the amount of native BPTI (Fig 4B). The native BPTI that ERp72 catalyzed was almost as much as that derived from spontaneous folding (Fig 4 A,C). ERp61 had a similar effect on the folding of N′ (Fig 4D). The native BPTI increased in the presence of ERp72 and ERp61 to almost the same extent as it increased by nonenzymatic reaction (Fig 4E). Although PDI dramatically increased the formation of native BPTI from N* compared with that obtained by a nonenzymatic reaction, ERp61 and ERp72 did not increase the amount of formation. PDI catalyzed the rearrangement of N′ to NSHSH and N* in the absence of redox reagents (Fig 5A). ERp61 and ERp72 catalyzed the rearrangement of N′ to NSHSH but did not catalyze the rearrangement of N′ to NSHSH or to N* in the absence of redox reagents, contrasting with PDI (Fig 5 B,C). The rate of accumulation of the native form in the presence of ERp61 or ERp72, however, was similar to that in the presence of PDI.
Fig 4.
Catalysis by ERp protein of the intramolecular rearrangement of the native 2-disulfide intermediate, N*, with both 2.0 mM GSH and 0.5 mM GSSG. The reactions with no enzyme (A), with PDI (B), with ERp72 (C), and with ERp61 (D) are indicated. (E) The kinetics of the native form:total BPTI ratio. Values were determined in the same manner as in Figure 2. PDI efficiently catalyzed the conversion of N* to N. ERp61 and ERp72 barely accelerated the conversion of N* to N in comparison with reaction in the absence of enzyme. Open circles, no enzyme; closed circles, 1.5 μM PDI; open squares, 1.5 μM ERp61; closed squares, 1.5 μM ERp72. GSSG, glutathione disulfide; PDI, protein disulfide isomerase; ER, endoplasmic reticulum
Fig 5.
Catalysis by ERp protein of the intramolecular rearrangement of N′ in the absence of redox reagents. The reactions with PDI (A), with ERp72 (B), and with ERp61 (C) are indicated. PDI, protein disulfide isomerase; ER, endoplasmic reticulum
ERp61 and ERp72 catalyzed the initial steps more efficiently than did PDI
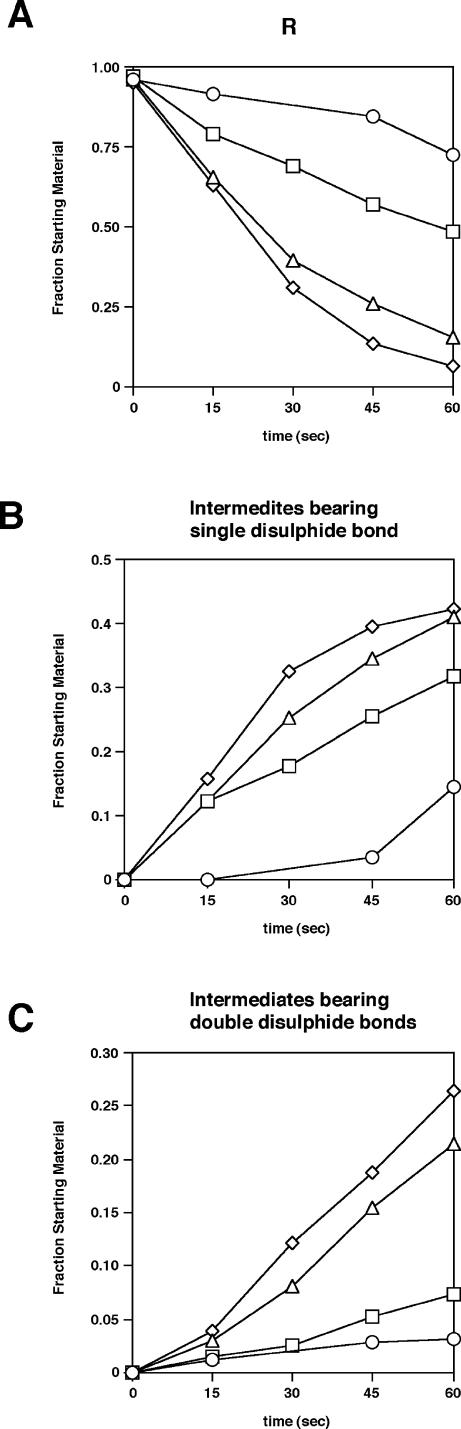
Although the reduced form was still detected at 2.5 minutes after the initiation of the reaction in the presence of PDI, it was hardly detected at the same incubation time in the presence of ERp61 and ERp72 (Fig 2). We compared the effects of each ERp protein on the disulfide bond formation in reduced BPTI in the initial steps, that is, the pathways from the reduced form (R), via intermediates bearing 1 disulfide bond ([30–51] and [5–55]) to intermediates bearing 2 disulfide bonds (N′, N*, and N′ [5-SSG, 55-SSG]). Seventeen percent (14% plus 3%) of the starting material showed the spontaneous formation of 1 or 2 disulfide bonds in the absence of ERp protein at 60 seconds after the initiation of the reaction (Fig 6). One and 2 disulfide bonds were formed in 32% and 7% of the starting material, respectively, formed in the presence of PDI at same incubation time (Fig 6). In contrast, the percentages of intermediates bearing 1 and 2 disulfide bonds increased up to 41% and 21%, respectively, in the presence of ERp61 at same incubation time (Fig 6). Similarly, the percentages of intermediates bearing 1 and 2 disulfide bonds increased up to 42% and 26%, respectively, in the presence of ERp72 at same incubation time (Fig 6).
Fig 6.
Conversion of fully reduced BPTI to intermediates bearing single or double disulfide bond. (A) Time course of decrease in the amount of starting material R, the fully reduced BPTI. (B) Time course of increase in the amount of intermediates bearing single disulfide bond [30–51] and [5–55]. (C) Time course of increase in the amount of intermediates bearing double disulfide bonds, N′, N*, and N′ [5-SSG, 55-SSG]. Values were determined in the same manner as in Figure 3. Circles, no enzyme; squares, PDI; triangles, ERp61; diamonds, ERp72. BPTI, bovine pancreatic trypsin inhibitor; PDI, protein disulfide isomerase; ER, endoplasmic reticulum
ERp72 and PDI cooperatively catalyzed BPTI folding
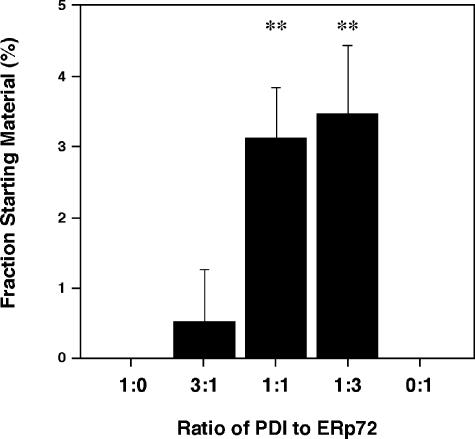
We investigated the effects of the coexistence of PDI and ERp72 on the BPTI-folding reaction (Fig 7). Fully folded BPTI was not observed in the reaction catalyzed by either PDI alone or ERp72 alone at 1 minute in the reaction (Fig 7). The combination of 1.125 μM PDI and 0.375 μM ERp72 folded 0.52% of the starting material to the native form at the same incubation time (Fig 7). The combination of 0.375 μM PDI and 1.125 μM ERp72 and that of 0.75 μM PDI and 0.75 μM ERp72 converted 3.5% and 3.1% of the starting material, respectively, to native BPTI at the same incubation time, and these combinations significantly increased the amount of native forms compared with PDI or ERp72 alone (Fig 7).
Fig 7.
Cooperation of PDI family proteins in the folding of BPTI. The ratio of fully folded BPTI to the starting material after the reduced BPTI was incubated for 1 minute. The reactions were initiated with either PDI alone, ERp72 alone, or 3 kinds of the combination of PDI and ERp72. Values were determined in the same manner as in Figure 2. Experiments were done 3 or 5 times in each group. Values are indicated as mean ± SD. 1:0, 1.5 μM PDI; 3:1, 1.125 μM PDI + 0.375 μM ERp72; 1:1, 0.75 μM PDI + 0.75 μM ERp72; 1:3, 0.375 μM PDI + 1.125 μM ERp72; 0:1, 1.5 μM ERp72. **P < 0.01. BPTI, bovine pancreatic trypsin inhibitor; PDI, protein disulfide isomerase; ER, endoplasmic reticulum
DISCUSSION
Native disulfide bonds are essential for the structure and function of many membrane and secretory proteins. For example, it is suggested that the modulation of redox potential of reactive thiols of skeletal muscle ryanodine receptor, a process in which disulfide formation is considered to be essential, is a general mechanism that controls the kinetics of the Ca2+ release (Xia et al 2000). Disulfide bond formation, reduction, and isomerization of membrane and secretory proteins in the ER of yeast are known to be catalyzed by PDI as well as Ero1p that reoxidizes PDI to regenerate the active site intrachain disulfide because oxidation of the substrate follows concomitantly the reduction of the PDI active site (Lyles and Gilbert 1991; Frand and Kaiser 1998, 1999; Pollard et al 1998). ERO1L-α and -β in mammalian cells, members of Ero1 family, are functionally equivalent to yeast Ero1p (Mezghrani et al 2001), and a growing family of oxidoreductases in mammalian ER are potential substrates for ERO1L-α and -β, including ERp61 and ERp72 (Jessop et al 2004). These indicate that ERp61 and ERp72 constitutively catalyze the formation, reduction, and isomerization of disulfide bonds in vivo.
In this study, we investigated the functional differences among PDI family proteins, PDI, ERp61, and ERp72, using BPTI as a model substrate that has been the best-characterized protein to analyze folding processes. Because PDI family proteins share 2 or 3 thioredoxinlike domains (Mazzarella et al 1990, 1994; Gilbert 1998), they have the capacity to catalyze disulfide bond formation and isomerization (Füllekrug et al 1994; Kozaki et al 1994; Rupp et al 1994; Bourdi et al 1995). ERp61 and ERp72 have a less efficient capacity for disulfide bond formation and isomerization compared with PDI (Füllekrug et al 1994; Kozaki et al 1994; Rupp et al 1994). Because the entire folding process of BPTI consists of several steps, we analyzed the individual steps separately.
PDI efficiently catalyzed the folding reaction from the fully reduced form (R) to the native form (N). In contrast, ERp61 and ERp72 accelerated the folding reaction from R to N more efficiently than spontaneous folding, but they converted only half of the molecules to the native form at 700 minutes. We addressed the functional differences between PDI and ERp61 or ERp72 at each step of BPTI folding. The study of oxidative folding of N′ (see Fig 3) revealed that PDI readily converted N′ to N, and partially to N*, indicating that PDI catalyzed the reduction of disulfide bond between residues 14 and 38 and the formation of disulfide bond between residues 5 and 55 and then recreated the disulfide bond between residues 14 and 38. ERp61 and ERp72 accelerated the conversion of N′ to N, but less efficiently than did PDI, and hardly converted N′ to N*. This result indicated that once the disulfide bond between residues 30 and 51 was formed, ERp61 and ERp72 were not able to reduce the disulfide bond between residues 14 and 38 and form the disulfide bond between residues 5 and 55.
The next study of oxidative folding of N* (see Fig 4) revealed that PDI also readily converted N* to N, indicating that PDI catalyzed the reduction of disulfide bond between residues 14 and 38 and the formation of disulfide bond between residues 30 and 51 and then recreated the disulfide bond between residues 14 and 38. ERp61 and ERp72 barely accelerated the conversion of N* to N in comparison with the reaction in the absence of enzyme. This result also indicated that once the disulfide bond between residues 5 and 55 was formed, ERp61 and ERp72 were not able to reduce the disulfide bond between residues 14 and 38 and form the disulfide bond residues 5 and 55. These results explained why only half of the molecules were converted to the native form and why an intermediate N* accumulated at 700 minutes after the initiation of the reaction. In both cases of conversions of N′ to N and N* to N, the reduction of disulfide bond between residues 14 and 38 was thought to be important to following disulfide bond formation. Given that N′ is essentially completely folded and that the rate-limiting step in the folding reaction is the loss of structure in N′, it is suggested that ERp61 and ERp72 are not able to distort the folded intermediates efficiently (Weissman and Kim 1991; Staley and Kim 1992).
The study of intramolecular rearrangement of disulfide bonds revealed that PDI accelerated the folding by rearrangement of N′ to N*, NSHSH (or to N). However, neither ERp61 nor ERp72 catalyzed the conversion of N′ to N* or to NSHSH. These results strongly suggested that ERp61 and ERp72 accelerated the folding of BPTI, not by rearranging the intramolecular disulfide bond of intermediates bearing 2 disulfide bonds but by catalyzing the direct oxidation of the third native disulfide bond. This was also supported by the results that ERp61 and ERp72 markedly increased the amount of N′ [5-SSG, 55-SSG], the direct oxidized specie of N′.
Because it is important for the rearrangement of N′ to N to lose structure, PDI accelerates the folding of BPTI by promoting both unfolding and disulfide bond rearrangements in structured intermediates (Weissman and Kim 1991, 1993). Because ERp61 and ERp72 did not readily convert intermediates, especially N*, to the native form, we consider that ERp61 and ERp72 do not possess the ability to unfold structured intermediates of BPTI. Alternatively, ERp61 may require the interaction with calnexin or calreticulin to unfold and rearrange in structured intermediates (Lindquist et al 2001; Antoniou et al 2002; Frickel et al 2002; Molinari et al 2002).
Although ERp61 and ERp72 yielded the native form in the lesser amount than did PDI, ERp61 and ERp72 catalyzed the disulfide bond formation more efficiently than did PDI. These results implied that the coexistence of PDI and ERp72 in the reaction catalyzed the folding of BPTI more efficiently than did PDI or ERp72 alone. We determined the most effective ratio of PDI to ERp72 in the formation of the fully folded BPTI by changing the ratio of PDI to ERp72 with the total amount of PDI family proteins being adjusted to 1.5 μM. The combination of 0.375 μM PDI and 1.125 μM ERp72 (PDI:ERp72 = 1:3) readily converted 3.5% of the reduced form to the native BPTI at 1 minute in the reaction. The combination of 0.75 μM PDI and 0.75 μM ERp72 (PDI:ERp72 = 1:1) folded 3.1% of the reduced form to the native form at the same incubation time. However, the combination of 1.125 μM PDI and 0.375 μM ERp72 (PDI:ERp72 = 3:1) hardly folded the reduced form to the native form at the same incubation time. These results indicated that PDI family proteins, when added together, cooperated with each other to fold BPTI by catalyzing the disulfide bond formation and isomerization and that the ratio of PDI to ERp72, rather than a simple coexistence, is critical for the effectiveness of BPTI folding. Although ERp61 or ERp72 had more efficient actions during the earlier half of the folding process of BPTI, PDI acted more efficiently during the later half of the folding process that is essential to the intramolecular rearrangements.
Whether or not the above-described ratio of PDI family proteins optimal for BPTI folding is suitable for folding of other proteins bearing disulfide bonds awaits further investigations.
The relative expression levels of PDI family proteins are different among cells from different tissues. Because cells from different tissues have different demands for secretory proteins to be synthesized, their requirements of PDI family proteins may vary from cell to cell accordingly, to form or isomerize disulfide bonds of secretory proteins.
Acknowledgments
This work was supported by Grant-in-Aid 10780452 and 15500250 for Scientific Research from Japan Society for the Promotion of Science.
REFERENCES
- Antoniou AN, Ford S, Alphey M, Osborne A, Elliott T, Powis SJ. The oxidoreductase ERp57 efficiently reduces partially folded in preference to fully folded MHC class I molecules. EMBO J. 2002;21:2655–2663. doi: 10.1093/emboj/21.11.2655.1460-2075(2002)021[2655:TOEERP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh S, Burns K, Andrin C, Michalak M. Interaction of calreticulin with protein disulfide isomerase. J Biol Chem. 1995;270:31338–31344. doi: 10.1074/jbc.270.52.31338.0021-9258(1995)270[31338:IOCWPD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bourdi M, Demady D, Martin JL, Jabbour SK, Martin BM, George JW, Pohl LR. cDNA cloning and baculovirus expression of the human liver endoplasmic reticulum P58: characterization as a protein disulfide isomerase isoform, but not as a protease or a carnitine acyltransferase. Arch Biochem Biophys. 1995;323:397–403. doi: 10.1006/abbi.1995.0060.0003-9861(1995)323[0397:CCABEO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Creighton TE. Protein folding coupled to disulphide bond formation. Biol Chem. 1997;378:731–744. doi: 10.1515/bchm.1997.378.8.731.1431-6730(1997)378[0731:PFCTDB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Frand AR, Cuozzo JW, Kaiser CA. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10:203–210. doi: 10.1016/s0962-8924(00)01745-1.0962-8924(2000)010[0203:PFPDBF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9.1097-2765(1998)001[0161:TEGOYI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7.1097-2765(1999)004[0469:EOPDII]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Freedman R, Bulleid N, Hawkins H, Paver J. Role of protein disulphide-isomerase in the expression of native proteins. Biochem Soc Symp. 1989;55:167–192.0067-8694(1989)055[0167:ROPDIT]2.0.CO;2 [PubMed] [Google Scholar]
- Freedman RB. Protein disulfide isomerase: multiple roles in the modification of nascent secretory proteins. Cell. 1989;57:1069–1072. doi: 10.1016/0092-8674(89)90043-3.0092-8674(1989)057[1069:PDIMRI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Freedman RB, Klappa P, Ruddock LW. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035.1469-221X(2002)003[0136:PDIESB]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel EM, Riek R, Jelesarov I, Helenius A, Wuthrich K, Ellgaard L. TROSY-NMR reveals interaction between ERp57 and the tip of the calreticulin P-domain. Proc Natl Acad Sci U S A. 2002;99:1954–1959. doi: 10.1073/pnas.042699099.0027-8424(2002)099[1954:TRIBEA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllekrug J, Sönnichsen B, Wünsch U, Arseven K, Van PN, Söling H-D, Mieskes G. CaBP1, a calcium binding protein of the thioredoxin family, is a resident KDEL protein of the ER and not of the intermediate compartment. J Cell Sci. 1994;107:2719–2727. doi: 10.1242/jcs.107.10.2719.0021-9533(1994)107[2719:CACBPO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0.0028-0836(1992)355[0033:PFITC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gilbert HF. Protein disulfide isomerase. Methods Enzymol. 1998;290:26–50. doi: 10.1016/s0076-6879(98)90005-2.0076-6879(1998)290[0026:PDI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3.0955-0674(1995)007[0523:QCITSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Iida K, Miyaishi O, Iwata Y, Kozaki K, Matsuyama M, Saga S. Distinct distribution of protein disulfide isomerase family proteins in rat tissues. J Histochem Cytochem. 1996;44:751–759. doi: 10.1177/44.7.8675996.0022-1554(1996)044[0751:DDOPDI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Jessop CE, Chakravarthi S, Watkins RH, Bulleid NJ. Oxidative protein folding in the mammalian endoplasmic reticulum. Biochem Soc Trans. 2004;32:655–658. doi: 10.1042/BST0320655.0300-5127(2004)032[0655:OPFITM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Johnson AE, van Waes MA. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799.1081-0706(1999)015[0799:TTADGA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kassenbrock CK, Kelly RB. Interaction of heavy chain binding protein (BiP/GRP78) with adenine nucleotides. EMBO J. 1989;8:1461–1467. doi: 10.1002/j.1460-2075.1989.tb03529.x.1460-2075(1989)008[1461:IOHCBP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki K, Miyaishi O, and Asai M. et al. 1994 Tissue distribution of ERp61 and association of its increased expression with IgG production in hybridoma cells. Exp Cell Res. 213:348–358. [DOI] [PubMed] [Google Scholar]
- Lee AS. The accumulation of three specific proteins related to glucose-regulated proteins in a temperature-sensitive hamster mutant cell line K12. J Cell Physiol. 1981;106:119–125. doi: 10.1002/jcp.1041060113.0021-9541(1981)106[0119:TAOTSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Mazzarella RA, Green M. Structure and assembly of the endoplasmic reticulum: biosynthesis and intracellular sorting of ERp61, ERp59, and ERp49, three protein components of murine endoplasmic reticulum. Arch Biochem Biophys. 1986;245:389–403. doi: 10.1016/0003-9861(86)90230-4.0003-9861(1986)245[0389:SAAOTE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lindquist JA, Hammerling GJ, Trowsdale J. ER60/ERp57 forms disulfide-bonded intermediates with MHC class I heavy chain. FASEB J. 2001;15:1448–1450. doi: 10.1096/fj.00-0720fje.0892-6638(2001)015[1448:EFDIWM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004.0006-2960(1991)030[0613:COTOFO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Marcus N, Shaffer D, Farrar P, Green M. Tissue distribution of three members of the murine protein disulfide isomerase (PDI) family. Biochim Biophys Acta. 1996;1309:253–260. doi: 10.1016/s0167-4781(96)00133-9.0006-3002(1996)1309[0253:TDOTMO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Martin J, Pumford N, LaRosa A, Martin B, Gonzaga H, Beaven M, Pohl L. A metabolite of halothane covalently binds to an endoplasmic reticulum protein that is highly homologous to phosphatidylinositol-specific phospholipase C-alpha but has no activity. Biochem Biophys Res Commun. 1991;178:679–685. doi: 10.1016/0006-291x(91)90161-y.0006-291X(1991)178[0679:AMOHCB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mazzarella RA, Marcus N, and Haugejörden SM. et al. 1994 Erp61 is GRP58, a stress-inducible luminal endoplasmic reticulum protein, but is devoid of phosphatidylinositide-specific phospholipase C activity. Arch Biochem Biophys. 308:454–460. [DOI] [PubMed] [Google Scholar]
- Mazzarella RA, Srinivasan M, Haugejorden SM, Green M. ERp72, an abundant luminal endoplasmic reticulum protein, contains three copies of the active site sequences of protein disulfide isomerase. J Biol Chem. 1990;265:1094–1101.0021-9258(1990)265[1094:EAALER]2.0.CO;2 [PubMed] [Google Scholar]
- Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288.1460-2075(2001)020[6288:MOOPFA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Galli C, Piccaluga V, Pieren M, Paganetti P. Sequential assistance of molecular chaperones and transient formation of covalent complexes during protein degradation from the ER. J Cell Biol. 2002;158:247–257. doi: 10.1083/jcb.200204122.0021-9525(2002)158[0247:SAOMCA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR, Rothman JE. Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem. 1987;56:829–852. doi: 10.1146/annurev.bi.56.070187.004145.0066-4154(1987)056[0829:BPTASB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0.1097-2765(1998)001[0171:EANAUP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Matlack KE, Plath K, Misselwitz B, Staeck O. Posttranslational protein translocation across the membrane of the endoplasmic reticulum. Biol Chem. 1999;380:1143–1150. doi: 10.1515/BC.1999.145.1431-6730(1999)380[1143:PPTATM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ruddon RW, Bedows E. Assisted protein folding. J Biol Chem. 1997;272:3125–3128. doi: 10.1074/jbc.272.6.3125.0021-9258(1997)272[3125:APF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rupp K, Birnbach U, Lundström J, Van PN, Söling H-D. Effects of CaBP2, the rat analog of ERp72, and of CaBP1 on the refolding of denatured reduced proteins. J Biol Chem. 1994;269:2501–2507.0021-9258(1994)269[2501:EOCTRA]2.0.CO;2 [PubMed] [Google Scholar]
- Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954.1471-0080(2002)003[0836:FATODB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Srivastava SP, Chen NQ, Liu YX, Holtzman JL. Purification and characterization of a new isozyme of thiol:protein-disulfide oxidoreductase from rat hepatic microsomes. Relationship of this isozyme to cytosolic phosphatidylinositol-specific phospholipase C form 1A. J Biol Chem. 1991;266:20337–20344.0021-9258(1991)266[20337:PACOAN]2.0.CO;2 [PubMed] [Google Scholar]
- Srivastava SP, Fuchs JA, Holtzman JL. The reported cDNA sequence for phospholipase C alpha encodes protein disulfide isomerase, isozyme Q-2 and not phospholipase-C. Biochem Biophys Res Commun. 1993;193:971–978. doi: 10.1006/bbrc.1993.1720.0006-291X(1993)193[0971:TRCSFP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Staley JP, Kim PS. Complete folding of bovine pancreatic trypsin inhibitor with only a single disulfide bond. Proc Natl Acad Sci U S A. 1992;89:1519–1523. doi: 10.1073/pnas.89.5.1519.0027-8424(1992)089[1519:CFOBPT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949.1081-0706(2003)019[0649:QCAPFI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Urade R, Nasu M, Moriyama T, Wada K, Kito M. Protein degradation by the phosphoinositide-specific phospholipase C-alpha family from rat liver endoplasmic reticulum. J Biol Chem. 1992;267:15152–15159.0021-9258(1992)267[15152:PDBTPP]2.0.CO;2 [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational proteintranslocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x.0076-6879(1983)096[0084:POMMFC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Weissman JS, Kim PS. Reexamination of the folding of BPTI: predominance of native intermediates. Science. 1991;253:1386–1393. doi: 10.1126/science.1716783.0193-4511(1991)253[1386:ROTFOB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Weissman JS, Kim PS. The pro region of BPTI facilitates folding. Cell. 1992;71:841–851. doi: 10.1016/0092-8674(92)90559-u.0092-8674(1992)071[0841:TPROBF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Weissman JS, Kim PS. Efficient catalysis of disulphide bond rearrangements by protein disulphide isomerease. Nature. 1993;365:185–188. doi: 10.1038/365185a0.0028-0836(1993)365[0185:ECODBR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Xia R, Stangler T, Abramson JJ. Skeletal muscle ryanodine receptor is a redox sensor with a well defined redox potential that is sensitive to channel modulators. J Biol Chem. 2000;275:36556–36561. doi: 10.1074/jbc.M007613200.0021-9258(2000)275[36556:SMRRIA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]