Abstract
One of the greatest challenges facing human geneticists is the identification and characterization of susceptibility genes for common complex multifactorial human diseases. This challenge is partly due to the limitations of parametric-statistical methods for detection of gene effects that are dependent solely or partially on interactions with other genes and with environmental exposures. We introduce multifactor-dimensionality reduction (MDR) as a method for reducing the dimensionality of multilocus information, to improve the identification of polymorphism combinations associated with disease risk. The MDR method is nonparametric (i.e., no hypothesis about the value of a statistical parameter is made), is model-free (i.e., it assumes no particular inheritance model), and is directly applicable to case-control and discordant-sib-pair studies. Using simulated case-control data, we demonstrate that MDR has reasonable power to identify interactions among two or more loci in relatively small samples. When it was applied to a sporadic breast cancer case-control data set, in the absence of any statistically significant independent main effects, MDR identified a statistically significant high-order interaction among four polymorphisms from three different estrogen-metabolism genes. To our knowledge, this is the first report of a four-locus interaction associated with a common complex multifactorial disease.
Introduction
The identification and characterization of susceptibility genes for common complex human diseases is one of the greatest challenges facing human geneticists. This challenge is partly due to the limitations of parametric-statistical methods (i.e., those in which a hypothesis about the value of a statistical parameter is made) for detection of gene effects that are dependent solely or partially on interactions with other genes (Templeton 2000) and with environmental exposures (Schlichting and Pigliucci 1998). For example, logistic regression is a commonly used method for modeling the relationship between discrete predictors, such as genotypes, and discrete clinical outcomes (Hosmer and Lemeshow 2000). However, logistic regression, like most parametric-statistical methods, is less practical for dealing with high-dimensional data. That is, when high-order interactions are modeled, there are many contingency-table cells that contain no observations (i.e., that are empty cells). This can lead to very large coefficient estimates and standard errors (Hosmer and Lemeshow 2000). One solution to this problem is to collect very large numbers of samples to allow robust estimation of interaction effects; however, the magnitudes of the samples that are often required incur prohibitive expense. An alternative solution is to develop new statistical and computational methods that have improved power to identify multilocus effects in relatively small samples.
To address this issue, we have developed a multifactor-dimensionality reduction (MDR) method for detecting and characterizing high-order gene-gene and gene-environment interactions in case-control and discordant-sib-pair studies with relatively small samples. The MDR method is inspired by the combinatorial-partitioning method (Nelson et al. 2001), a data-reduction method for the exploratory analysis of quantitative traits. With MDR, multilocus genotypes are pooled into high-risk and low-risk groups, effectively reducing the genotype predictors from n dimensions to one dimension. The new, one-dimensional multilocus-genotype variable is evaluated for its ability to classify and predict disease status through cross-validation and permutation testing. The MDR method is model free—in that it does not assume any particular genetic model—and is nonparametric—in that it does not estimate any parameters. We first evaluate the MDR method by using simulated multilocus data with epistatic effects, and we then apply it to identification of multiple single-nucleotide polymorphisms associated with sporadic breast cancer.
Breast cancer is generally considered a complex disease, since its most common form—sporadic breast cancer—is undoubtedly due to multiple unknown etiologies. This is in contrast to the less common form—familial breast cancer, which is attributed to single-gene abnormalities (e.g., BRCA1 [MIM 113705] and BRCA2 [MIM 600185]). Although the causes of sporadic breast cancer remain undetermined, there is substantial experimental, epidemiological, and clinical evidence that estrogens influence breast cancer risk (Clemons and Goss 2001). In fact, recent evidence indicates that the oxidative metabolism of estrogens to catechol estrogens and to estrogen quinones can cause mutagenic DNA lesions (Yager and Liehr 1996; Cavalieri et al. 1997; Parl 2000). Consequently, catechol estrogen and estrogen quinones have been implicated in mammary carcinogenesis. The catechol-estrogen pathway is regulated by catechol-O-methyltransferase (COMT), by cytochromes P450 1A1 and P450 1B1 (CYP1A1 and CYP1B1, respectively), and by glutathione S-transferases M1 and T1 (GSTM1 and GSTT1, respectively). Each of the genes encoding these enzymes contains functional polymorphisms that result in different concentrations of catechol-estrogen metabolites (Seidegard et al. 1988; Hayashi et al. 1991; Wiencke et al. 1995; Cascorbi et al. 1996; Lachman et al. 1996; Persson et al. 1997; Syvanen et al. 1997; Bailey et al. 1998a; Stoilov et al. 1998; Hanna et al. 2000). We hypothesize that interactions between polymorphisms of these genes may have a synergistic, or nonadditive, effect on the pathogenesis of breast cancer and, thereby, may explain differences in breast cancer risk. Application of MDR to a sporadic breast cancer case-control data set, in the absence of any statistically significant independent main effects, identified a statistically significant high-order interaction among four polymorphisms from three different estrogen-metabolism genes—COMT (MIM 116790), CYP1B1 (MIM 601771), and CYP1A1 (MIM 108330).
Subjects and Methods
MDR
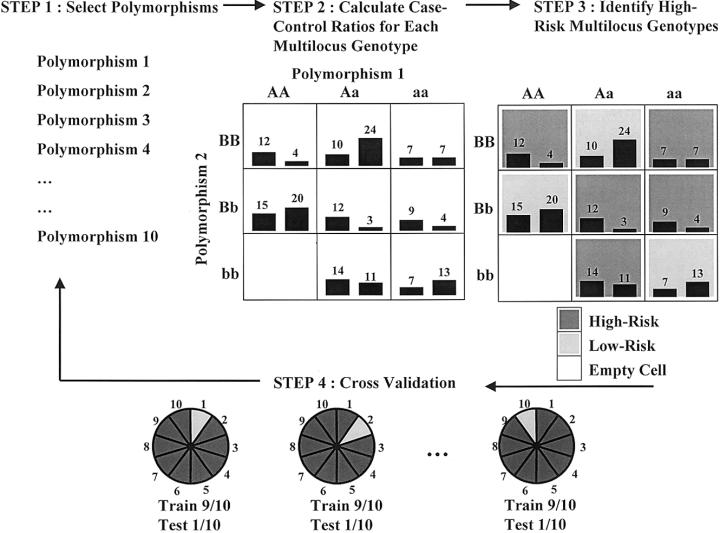
Figure 1 illustrates the four general steps involved in implementing the MDR method for case-control studies. The same procedure is equally applicable to discordant-sib-pair studies. In step 1, a set of n genetic and/or discrete environmental factors is selected from the pool of all factors. In step 2, the n factors and their possible multifactor classes or cells are represented in n-dimensional space; for example, for two loci with three genotypes each, there are nine two-locus–genotype combinations. Then, the ratio of the number of cases (or affected sibs) to the number of controls (or unaffected sibs) is estimated within each multifactor class. In step 3, each multifactor cell in n-dimensional space is labeled either as “high-risk,” if the cases:controls ratio meets or exceeds some threshold (e.g., ⩾1.0), or as “low-risk,” if that threshold is not exceeded. In this way, a model for both cases and controls (or for affected and unaffected sibs) is formed by pooling high-risk cells into one group and low-risk cells into another group. This reduces the n-dimensional model to a one-dimensional model (i.e., having one variable with two multifactor classes—high risk and low risk). In this initial implementation of MDR, balanced case-control studies are required. In step 4, the prediction error of each model is estimated by 10-fold cross-validation. Here, the data (i.e., subjects) are randomly divided into 10 equal parts. The MDR model is developed for each possible 9/10 of the subjects and then is used to make predictions about the disease status of each possible 1/10 of the subjects excluded. The proportion of subjects for which an incorrect prediction was made is an estimation of the prediction error. To reduce the possibility of poor estimates of the prediction error that are due to chance divisions of the data set, the 10-fold cross-validation is repeated 10 times, and the prediction errors are averaged.
Figure 1.
Summary of steps involved in implementation of the MDR method: a set of n genetic and/or discrete environmental factors is selected; the n factors and their possible multifactor classes or cells are represented in n-dimensional space; each multifactor cell in n-dimensional space is labeled as either “high-risk” or “low-risk”; and the prediction error of each model is estimated. For each multifactor combination, hypothetical distributions of cases (left bars in boxes) and of controls (right bars in boxes) are shown.
For studies with more than two factors, the four steps of the MDR method are repeated for each possible combination, if computationally feasible. If the number of combinations to be evaluated exceeds computational feasibility, machine learning methods, such as parallel genetic algorithms (Cantú-Paz 2000), must be employed. Among all of the two-factor combinations, a single model that maximizes the cases:controls ratio of the high-risk group is selected. This two-locus model will have the minimum classification error among the two-locus models. Single best multifactor models are also selected from among the models for each of the three- to n-factor combinations. Among this set of best multifactor models, the combination of loci and/or discrete environmental factors that minimizes the prediction error is selected. Thus, the classification errors and the prediction errors estimated by 10-fold cross-validation are used to select the final multifactor model. Hypothesis testing for this final model can then be performed by evaluating the consistency of the model across cross-validation data sets—that is, how many times the same MDR model is identified in each possible 9/10 of the subjects. The reasoning is that a true signal (i.e., association) should be present in the data regardless of how they are divided. We determined statistical significance by comparing the average cross-validation consistency from the observed data to the distribution of average consistencies under the null hypothesis of no associations derived empirically from 1,000 permutations. The null hypothesis was rejected when the upper-tail Monte Carlo P value derived from the permutation test was ⩽.05.
Data Simulation
To evaluate the MDR method, we simulated four sets of 50 replicates of 200 cases and 200 controls, using four different multilocus epistasis models. This number of replicates was selected to be large enough to provide validation of the method and to be small enough to allow exhaustive computational searches of all possible multilocus models. Unrelated subjects and genotypes for 10 unlinked biallelic loci were simulated by the Genometric Analysis Simulation Package (Wilson et al. 1996). Allele frequencies for each of the 10 loci were selected to match those in the sporadic–breast cancer case-control sample. Hardy-Weinberg equilibrium and linkage equilibrium were assumed. For the first model, we simulated a two-locus interaction effect, using penetrance functions P(D|AAbb) = .2, P(D|AaBb) = .2, P(D|aaBB) = .2, and P(D|others) = 0, where D is disease and A, a, B, and b represent the alleles for the disease-susceptibility loci. This is a well-characterized model for epistasis, in which disease risk is dependent on whether two deleterious alleles and two normal alleles are present, from either one locus or both loci (Frankel and Schork 1996; Li and Reich 2000). As described by Frankel and Schork (1996) and by Li and Reich (2000), the independent main effects for the loci in this model are small. We extended this two-locus epistasis model to three-locus, four-locus, and five-locus epistasis models by adding corresponding homozygous or heterozygous genotypes to the aforementioned penetrance functions. For example, for the three-locus epistasis model, we used penetrance functions P(D|AAbbcc) = .2, P(D|AaBbcc) = .2, P(D|aaBBcc) = .2, P(D|aaBbCc) = .2, P(D|AabbCc) = .2, and P(D|aabbCC) = .2. Thus, of the 10 total simulated loci, there were 2, 3, 4, or 5 functional epistatic loci and up to 8 nonfunctional loci.
Sporadic–Breast Cancer Data
This study is based on 200 white women with sporadic primary invasive breast cancer who were treated at Vanderbilt University Medical Center during 1982–96. Informed consent for this study was obtained from all study subjects, in accordance with the requirements of the Institutional Review Board of Vanderbilt University Medical School. Breast cancer was classified as either sporadic or familial, on the basis of family history as determined by patient questionnaire: patients with either at least one first-degree relative with breast cancer or at least two second-degree relatives with breast cancer were considered to have familial breast cancer; patients not fulfilling these criteria were considered to have sporadic breast cancer. Patients with sporadic breast cancer were frequency age-matched to control patients at Vanderbilt University Medical Center who had been hospitalized for various acute and chronic illnesses. Reasons for exclusion of controls included breast cancer or other forms of malignancy, as well as family history of breast cancer.
DNA was isolated from all samples by use of a DNA extraction kit (Gentra). Because their enzyme products interact in the metabolism of estrogens to catechol estrogens and to estrogen quinones, our analysis focused on the genes COMT (MIM 116790), on chromosome 22q11.2; CYP1A1 (MIM 108330), on chromosome 15q22-qter; CYP1B1 (MIM 601771), on chromosome 2p21-22; GSTM1 (MIM 138350), on chromosome 1p13.3; and GSTT1 (MIM 600436), on chromosome 22q11.2. COMT and GSTT1 are ∼4 Mb apart on chromosome 22q11.2. Table 1 summarizes the polymorphisms, in these genes, that we analyzed by PCR and restriction-endonuclease digestion. Genotype frequencies have been previously reported by our group (Bailey et al. 1998a, 1998b; Parl 2000) and by others (Lavigne et al. 1997; Millikan et al. 1998; Thompson et al. 1998). The specific primers and amplification conditions and the subsequent restriction-endonuclease analysis for CYP1A1, CYP1B1, GSTM1, and GSTT1 have been described elsewhere (Bailey et al. 1998a, 1998b). COMT was amplified with primers C1 (5′-GCC GCC ATC ACC CAG CGG ATG GTG GAT TTC GCT GTC) and C2 (5′-GTT TTC AGT GAA CGT GGT GTG). Each PCR contained internal controls for the respective gene, and random retesting of ∼5% of the samples yielded 100% reproducibility.
Table 1.
Enzyme Genotype Analysis by PCR and Restriction-Endonuclease Digestion
|
Polymorphism |
Genotype Frequencya(%) |
||||||
| Enzyme | Nucleotide | Codon | Primers | Endonuclease | w/w | w/p | p/p |
| COMT | 1947G→A | 158Val→Met | C1, C2 | BspHI | 25 | 51 | 24 |
| CYP1A1: | |||||||
| m1 | T6235T→C | 3′ UTR | A3, A4b | MspI | 82 | 15 | 3 |
| m2 | 4887C→A | 461Thr→Asn | A1, A4b | BsaI | 92 | 7 | 1 |
| m4 | 4889A→G | 462Ile→Val | A1, A2b | BsrDI | 92 | 8 | 0 |
| CYP1B1: | |||||||
| Codon 48 | 143C→G | 48Arg→Gly | B1, B2c | RsrII | 51 | 40 | 9 |
| Codon 119 | 355G→T | 119Ala→Ser | B1, B2c | NgoMIV | 51 | 40 | 9 |
| Codon 432 | 1294G→C | 432Val→Leu | B3, B4c | Eco57I | 12 | 58 | 30 |
| Codon 453 | 1358A→G | 453Asn→Ser | B3, B4c | Cac8I | 68 | 30 | 2 |
| GSTM1 | Deletion | Loss of enzyme | M1, M2b | … | 57d | 43 | |
| GSTT1 | Deletion | Loss of enzyme | T1, T2b | … | 79d | 21 | |
Data Analysis
Prior to application of MDR to the sporadic–breast cancer data set, the method was evaluated by use of the simulated multilocus data sets. For each of the 50 replicates generated by each of the four multilocus epistasis models, we applied the MDR algorithm as described in the subsection “MDR,” with a threshold cases:controls ratio of at least 1:1. This threshold was selected so that multilocus-genotype combinations would be considered high-risk if the number of cases with that particular combination either was equal to or exceeded the number of controls; whether more-stringent thresholds improve the results will be the focus of future studies. An exhaustive search of all possible two- to nine-locus models was performed. The 10-locus model was not evaluated, since there is only one such model and since its cross-validation consistency is always 10. On validation of the method, MDR was then applied to the sporadic–breast cancer data set, with the same threshold cases:controls ratio, at least 1:1. An exhaustive search of all possible two- to nine-locus models was again performed.
Results
Application of MDR to Simulated Data
Table 2 summarizes the means and the standard errors of the means (SEMs), of both the cross-validation consistency and the prediction error, obtained from the MDR analysis of each group of 50 simulated data sets for each gene-gene interaction model and each number of loci evaluated. For the particular multilocus models that contain the correct two, three, four, or five genes, for each group of 50 simulated data sets, the mean prediction error was minimum, and the mean cross-validation consistency was maximum. Additionally, the SEM of the prediction error and of the cross-validation consistency was minimum at the correct multilocus model. For example, in the case in which a three-locus epistasis model was used to simulate the data sets, the mean ± SEM prediction error was minimum for the three-locus model, at 12%±0.22%. The two-locus models had a mean ± SEM prediction error of 21.91%±0.33%), whereas the four-locus model had a mean ± SEM prediction error of 12.37%±0.24%. The mean prediction error for the four-locus model was much closer to that of the three-locus model, because these models contained the correct three functional loci as well as a false-positive locus, whereas the two-locus models were missing one of the functional loci. Selecting the smaller three-locus model with the lower mean prediction error is consistent with statistical parsimony (i.e., smaller models are better because they are easier to interpret). For the three-locus models in this example, the cross-validation consistency was always 10.00; that is, the same three-locus model was found in each possible 9/10 of the subjects. These results suggest that, for this particular epistasis model, the cross-validation strategy is a reasonable approach to the identification of the correct multilocus model. Furthermore, the threshold cases:controls ratio of at least 1:1 was reasonable for this epistasis model.
Table 2.
Summary of Simulation Results[Note]
|
Mean ± SEM |
||
| No. of Locia | Cross-Validation Consistency | Prediction Error |
| Model 2: | ||
| 2 | 9.86 ± .08 | 14.99 ± .24 |
| 3 | 7.41 ± .21 | 15.58 ± .26 |
| 4 | 6.01 ± .22 | 16.49 ± .29 |
| 5 | 5.56 ± .24 | 19.03 ± .38 |
| 6 | 6.52 ± .34 | 23.23 ± .53 |
| 7 | 6.94 ± .26 | 24.49 ± .62 |
| 8 | 7.90 ± .29 | 25.02 ± .73 |
| 9 | 8.03 ± .23 | 25.40 ± .73 |
| Model 3: | ||
| 2 | 9.20 ± .17 | 21.91 ± .33 |
| 3 | 10.00 ± .00 | 12.00 ± .22 |
| 4 | 9.27 ± .13 | 12.37 ± .24 |
| 5 | 6.28 ± .21 | 13.90 ± .28 |
| 6 | 5.86 ± .25 | 15.57 ± .32 |
| 7 | 6.26 ± .29 | 17.75 ± .43 |
| 8 | 7.68 ± .28 | 19.39 ± .47 |
| 9 | 7.99 ± .25 | 19.93 ± .50 |
| Model 4: | ||
| 2 | 8.40 ± .26 | 19.15 ± .35 |
| 3 | 8.79 ± .20 | 10.20 ± .23 |
| 4 | 10.00 ± .00 | 5.68 ± .17 |
| 5 | 9.32 ± .12 | 6.02 ± .19 |
| 6 | 7.74 ± .16 | 6.88 ± .22 |
| 7 | 7.01 ± .22 | 7.73 ± .26 |
| 8 | 7.04 ± .24 | 8.64 ± .31 |
| 9 | 7.79 ± .24 | 9.46 ± .34 |
| Model 5: | ||
| 2 | 9.01 ± .20 | 15.33 ± .28 |
| 3 | 8.37 ± .25 | 8.54 ± .24 |
| 4 | 8.16 ± .25 | 5.17 ± .20 |
| 5 | 9.99 ± .01 | 2.95 ± .11 |
| 6 | 9.52 ± .12 | 3.17 ± .14 |
| 7 | 9.13 ± .16 | 3.66 ± .17 |
| 8 | 8.74 ± .17 | 4.17 ± .19 |
| 9 | 9.00 ± .14 | 4.60 ± .18 |
Note.— For each simulation model, the multilocus model with maximum mean ± SEM cross-validation consistency and minimum mean ± SEM prediction error is indicated in boldface italic type.
Model number is based on the number of epistatic genes in each simulation model.
The Monte Carlo P values for each of the correctly identified models were all <.001. The estimated power to identify the correct multilocus model was 78% for the two-locus model, 82% for the three-locus model, 94% for the four-locus model, and 90% for the five-locus model. It is interesting that the power to identify the correct multilocus model tends to increase as higher-order interactions are modeled. This may be a real phenomenon, or it may be due to the fact that fewer nonfunctional loci of the 10 that were simulated were present; this will require further investigation. These results suggest that, for this particular epistasis model, the MDR method has reasonable power to identify high-order gene-gene interactions in a sample of 200 cases and 200 controls.
Application of MDR to Breast Cancer Data
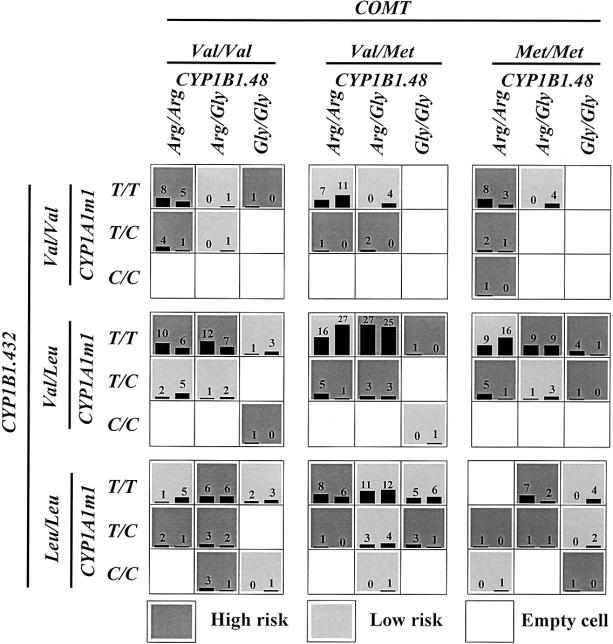
Table 3 summarizes the cross-validation consistency and the prediction error obtained from MDR analysis of the sporadic–breast cancer case-control data set, for each number of loci evaluated. One four-locus model had a minimum prediction error of 46.73 and a maximum cross-validation consistency of 9.8 that was significant at the .001 level, as determined empirically by permutation testing. Thus, under the null hypothesis of no association, it is highly unlikely that a cross-validation consistency ⩾9.8 will be observed for this four-locus model. The four-locus model included the polymorphisms of COMT, CYP1A1m1, CYP1B1 codon 48, and CYP1B1 codon 432. Figure 2 summarizes the four-locus–genotype combinations associated with high risk and with low risk, along with the corresponding distribution of cases and of controls, for each multilocus-genotype combination. Note that the patterns of high-risk and low-risk cells differ across each of the different multilocus dimensions. This is evidence of epistasis, or gene-gene interaction; that is, the influence that each genotype at a particular locus has on disease risk is dependent on the genotypes at each of the other three loci. Previous analysis of this data set, by logistic regression, revealed no statistically significant evidence of independent main effects of any of the 10 polymorphisms (Bailey et al. 1998a, 1998b; authors' unpublished data).
Table 3.
Summary of Results for Breast Cancer[Note]
| No. of Loci | Cross-Validation Consistency | Prediction Error |
| 2 | 7.00 | 51.06 |
| 3 | 4.17 | 51.35 |
| 4 | 9.80a | 46.73 |
| 5 | 4.71 | 50.26 |
| 6 | 5.00 | 48.61 |
| 7 | 8.60 | 47.15 |
| 8 | 8.20 | 52.55 |
| 9 | 7.10 | 53.40 |
Note.— The multilocus model with maximum cross-validation consistency and minimum prediction error is indicated in boldface italic type.
P<.001.
Figure 2.
Summary of four-locus genotype combinations associated with high risk and with low risk for sporadic breast cancer, along with the corresponding distribution of cases (left bars in boxes) and of controls (right bars in boxes), for each multilocus-genotype combination. Note that the patterns of high-risk and low-risk cells differ across each of the different multilocus dimensions. This is evidence of epistasis, or gene-gene interaction.
Discussion
We have introduced MDR as a method for reducing the dimensionality of multilocus information, to improve identification of combinations of polymorphisms associated with the risk for common complex multifactorial diseases. The development of MDR was motivated by the limitations of the generalized linear model for detection and characterization of gene-gene (Templeton 2000) and gene-environment (Schlichting and Pigliucci 1998) interactions and by the success of data-reduction methods for quantitative traits (Nelson et al. 2001). Using simulated data, we demonstrated the applicability of MDR for identification of genes whose effects are primarily through interaction. We then applied MDR to identify gene-gene interaction effects on risk for sporadic breast cancer.
Breast cancer is generally considered a multifactorial disease with estrogens as one of the principal factors. We therefore applied MDR to a set of genes (i.e., COMT, CYP1A1, CYP1B1, GSTM1, and GSTT1) whose protein products interact as enzymes in the metabolism of estrogens in breast tissue. Several studies have examined the breast cancer risk associated with individual genotypes of each of these enzymes (Rebbeck et al. 1994; Ambrosone et al. 1995; Lavigne et al. 1997; Bailey et al. 1998a; Millikan et al. 1998; Thompson et al. 1998). Not surprisingly, the results have been inconsistent and even contradictory. That is, if a single gene in the estrogen-metabolism pathway were solely responsible for breast cancer, then the malignancy would likely present as familial breast cancer, and the gene would be identified by linkage analysis, as in the case of BRCA1 and BRCA2. Studies of two or three genotypes in combination have also yielded inconsistent results. For example, we examined CYP1A1, GSTM1, and GSTT1 polymorphisms in a case-control study of 328 white and 108 African American women, using multiple logistic-regression analysis (Bailey et al. 1998b). None of the enzyme genotypes—individually or combined—were associated with an increased risk for breast cancer. However, we did not include COMT and CYP1B1 in the analysis, because their roles in the catechol-estrogen pathway and/or their various polymorphisms were only recently elucidated (Yager and Liehr 1996; Cavalieri et al. 1997; Bailey et al. 1998a; Stoilov et al. 1998; Parl 2000). Because of their clearly defined functional interactions in the catechol-estrogen pathway, it is essential to consider the combined effect of all these enzymes. In this article, we have demonstrated that the MDR applied to 10 single-nucleotide polymorphisms in COMT, CYP1A1, CYP1B1, GSTM1, and GSTT1 identifies a four-locus interaction that is significantly associated with risk for sporadic breast cancer. To our knowledge, this is the first report of a four-locus interaction associated with a common complex multifactorial disease.
Many groups, including our own, have reported that breast cancer risk is influenced by several nongenetic hormonal factors, such as age at menarche, and by age at menopause, body-mass index, reproductive history, lactation history, and use of exogenous estrogen in the form of either oral contraceptives or hormone-replacement therapy (Kelsey and Berkowitz 1988; Dupont et al. 1989; Harris et al. 1992; Kelsey et al. 1993; Collaborative Group on Hormonal Factors in Breast Cancer 1996, 1997). Although these factors allow prediction of a relative risk for a given population, they are not very helpful to individual women. As defined by the MDR, the determination of a woman’s genotype may add another dimension to the assessment of overall breast cancer risk. However, it is obvious that there is also an interaction between genotype risk factors and traditional hormonal risk factors. For example, obesity has been related both to the concentration of endogenous estrogen and to breast cancer risk. Several studies have demonstrated that obese postmenopausal women have an increased risk for breast cancer, compared to age-matched nonobese postmenopausal women (Harris et al. 1992; Yong et al. 1996). The elevated risk has been attributed to higher levels of circulating estrogens secondary to increased conversion, in adipose tissue, of androgen to estrogen. Several studies have demonstrated significantly higher serum-estradiol concentrations in obese postmenopausal women than in their nonobese counterparts (MacDonald et al. 1978; Moore et al. 1987; Potischman et al. 1996). Thus, any effect that COMT, CYP1A1, CYP1B1, GSTM1, and GSTT1 may have on estrogen metabolism may be affected by the concentration of estradiol. Consequently, our present analysis of genetic factors is limited by lack of consideration of these traditional hormonal risk factors.
The Advantages of MDR
The primary advantage of MDR is that it facilitates the simultaneous detection and characterization of multiple genetic loci associated with a discrete clinical endpoint. This is accomplished by reducing the dimensionality of the multilocus data. In essence, genotypes from multiple loci and/or discrete environmental classes are pooled into high-risk and low-risk groups, depending on whether they are more common in affected or in unaffected subjects. This new multilocus-genotype encoding reduces the dimensionality to one. For the simulated data, the mean cross-validation consistency was always maximized, and the mean prediction error was always minimized, at the correct multilocus model.
Another important advantage of MDR is that it is nonparametric. This is an important difference versus traditional parametric-statistical methods, which rely on the generalized linear model. For example, in logistic regression, as each additional main effect is included in the model, the number of possible interaction terms grows exponentially. Having too many independent variables in relation to the number of observed outcome events is a well-recognized problem (Concato et al. 1993). Simulation studies by Peduzzi et al. (1996) suggest that having fewer than 10 outcome events per independent variable can lead to biased estimates of the regression coefficients and to an increase in type 1 and type 2 errors. For example, with two outcome events per independent variable, more than one-third of the estimated regression coefficients differed from the true parameter value by a magnitude of 2 (Peduzzi et al. 1996). Hosmer and Lemeshow (2000) suggest that logistic-regression models should contain no more than P+1⩽min(n1,n0)/10 parameters, where n1 is the number of events of type 1 and n0 is the number of events of type 0. For the 200 cases and the 200 controls evaluated in the present study, this formula suggests that no more than 19 parameters should be estimated in a logistic-regression model. In a logistic-regression model, how many parameters must be estimated to identify interactions among the 10 estrogen-metabolism–gene polymorphisms? The number of orthogonal-regression terms needed to describe the interactions among a subset, k, of n biallelic loci is (n choose k)×2k (Wade 2000). Thus, for 10 genes, we would need 20 parameters to model the main effects (assuming two dummy variables per biallelic locus), 180 parameters to model the two-way interactions, 1,920 parameters to model the three-way interactions, 3,360 parameters to model the four-way interactions, and so forth. Thus, fitting a full model with all interaction terms and then using backward elimination to derive a parsimonious model would not be possible. The MDR method avoids the problems associated with the use of parametric statistics to model high-order interactions.
A third advantage of MDR is that it assumes no particular genetic model (i.e., it is model free); that is, no mode of inheritance needs to be specified. This is important for diseases, such as sporadic breast cancer, in which the mode of inheritance is unknown and likely very complex. In its current form, MDR can be directly applied to case-control and discordant-sib-pair studies. Extension to other family-based control study designs, such as those using trios, should also be possible.
A fourth advantage of MDR is that false-positive results due to multiple testing are minimized. This is primarily due to the cross-validation strategy used to select optimal models. Data-reduction and pattern-recognition methods are good for identification of complex relationships among data, even when those relationships are due to either chance or false-positive variations. However, the real test of any method is its ability to make predictions in independent data (Ripley 1996). Cross-validation divides the data into 10 equal parts, allowing 9/10 of the data to be used to develop a model and the independent 1/10 of the data to be used to evaluate the predictive ability of the model. Optimal models are selected solely on the basis of their ability to make predictions with regard to independent data. Only when a final predictive model has been selected is the null hypothesis of no association tested via permutation testing. It is this combined cross-validation–testing/permutation-testing method that minimizes false-positives due to multiple examinations of the data.
The Disadvantages and Limitations of MDR
Although MDR overcomes some of the limitations of the generalized linear model, there are three important disadvantages. First, MDR can be computationally intensive, especially when more than 10 polymorphisms need to be evaluated. A genome scan with hundreds to thousands of polymorphisms requires robust machine learning algorithms, since all of the possible multilocus combinations cannot be exhaustively searched. This is, however, a limitation of any multilocus method that does not first condition on a particular locus having an independent main effect (e.g., stepwise logistic regression). Second, MDR models can be difficult to interpret. This is illustrated clearly in the four-locus model in figure 2. There are no obvious trends or patterns in the distribution of high-risk and low-risk groupings across the four-dimensional genotype space; for example, a consistent trend of high-risk or low-risk cells across a series of rows or of columns may indicate that a particular locus has a main effect. The lack of such trends in the four-locus model for breast cancer is indicative of epistasis; that is, the influence of each genotype on disease risk appears to be dependent on the genotypes at each of the other loci. Sorting out the nature of the interactions in four-dimensional space to infer function remains an interpretive challenge. Third, in its current form, MDR can be applied only to case-control studies that are balanced (i.e., that have the same number of cases and of controls). This limitation will be addressed in future studies (see the following subsection, “Future Studies”).
Another limitation of MDR is its ability to make predictions for independent data sets when the dimensionality of the best model is relatively high and the sample is relatively small. High dimensionality and a small sample lead to many multifactor cells with either missing data or singleton data. This is not a problem for estimation of the classification error and evaluation of the cross-validation consistency, but it is a problem for estimation of the prediction error. For example, if there were one observation for each multifactor cell in n-dimensional space, then, during cross-validation, that one observation will end up in either the training data used to estimate the classification error or the test data used to estimate the prediction error but not in both. If the observation ends up in the test data, there will be, from the training data, no model (i.e., there will be an empty cell) to make a prediction. This greatly limits the number of observations for which predictions can be made in the test set and ultimately impacts the SEM of the prediction error. Proposed future studies will address this limitation (see the following subsection, “Future Studies”).
Future Studies
The MDR is a powerful alternative to traditional parametric statistics such as logistic regression. We have demonstrated the MDR's ability to identify high-order (i.e., more than two) gene-gene interactions in relatively small simulated and real data sets. Although MDR addresses some of the limitations of the generalized linear model, there are several ways in which the method can be improved.
First, if MDR is going to be used for genome scans with hundreds to thousands of single-nucleotide polymorphisms, then it will be necessary to develop machine learning strategies to optimize the selection of polymorphisms to be modeled, since an exhaustive search of all possible combinations will not be possible. We are currently exploring the use of parallel genetic algorithms (Cantú-Paz 2000) as a robust machine learning approach.
Second, it will be important to improve MDR's predictive ability in the higher dimensions. We are currently exploring several strategies to improve the estimation of the prediction error. The first strategy uses a nearest-neighbor method to determine whether an empty cell should be classified as high risk or as low risk; for example, if the majority of multilocus-genotype combinations within one step in n-dimensional space are classified as high risk, then the empty cell is also classified as high risk. The second strategy projects either a high risk or a low risk classification for an empty cell in a lower dimension; for example, the locus with the least-frequent genotype might be removed from the model, and risk could then be determined from the equivalent genotypes in a lower dimension. These strategies will be compared to determine whether either improves the estimation of the prediction error when empty cells are present.
Third, it will be important to modify MDR for the analysis of unbalanced case-control studies. We are currently exploring several different weighting schemes for the case-control ratio that account for whether the total number of cases or the total number of controls is greater. Finally, simulation studies will be needed to determine the strengths and the weaknesses of MDR in the presence of genotyping errors, phenocopies, genetic heterogeneity, and other phenomena that complicate the identification and characterization of functional polymorphisms. We anticipate that data-reduction methods such as MDR will be invaluable for the identification and characterization of high-order gene-gene and high-order gene-environment interactions, when few degrees of freedom are available for parametric-statistical estimation of interaction effects.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant RO1 CA/ES83752 and by generous funds from the Vanderbilt-Ingram Cancer Center and from the Vanderbilt University Medical School. M.D.R. was supported by NIH training grant T32 CA78136. We thank Dr. Scott Williams for critical reading of the manuscript, and we thank two anonymous reviewers for very helpful comments and suggestions.
Electronic-Database Information
Accession numbers and the URL for data in this article are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for COMT [MIM 116790], CYP1A1 [MIM 108330], CYP1B1 [MIM 601771], GSTM1 [MIM 138350], and GSTT1 [MIM 600436])
References
- Ambrosone CB, Freudenheim JL, Graham S, Marshall JR, Vena JE, Brasure JR, Laughlin R, et al (1995) Cytochrome P4501A1 and glutathione S-transferase (M1) genetic polymorphisms and postmenopausal breast cancer risk. Cancer Res 55:3483–3485 [PubMed] [Google Scholar]
- Bailey LR, Roodi N, Dupont WD, Parl FF (1998a) Association of cytochrome P450 1B1 (CYP1B1) polymorphism with steroid receptor status in breast cancer. Cancer Res 58:5038–5041 (erratum: Cancer Res 59:1388 [1999]) [PubMed] [Google Scholar]
- Bailey LR, Roodi N, Verrier CS, Yee CJ, Dupont WD, Parl FF (1998b) Breast cancer and CYP1A1, GSTM1, and GSTT1 polymorphisms: evidence of a lack of association in Caucasians and African Americans. Cancer Res 58:65–70 [PubMed] [Google Scholar]
- Cantú-Paz E (2000) Efficient and accurate parallel genetic algorithms. Kluwer Academic, Boston [Google Scholar]
- Cascorbi I, Brockmoller J, Roots I (1996) A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res 56:4965–4969 [PubMed] [Google Scholar]
- Cavalieri EL, Stack DE, Devanesan PD, Todorvic R, Dwivedy I, Higginbotham S, Johansson SL, et al (1997) Molecular origin of cancer: catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci USA 94:10937–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons M, Goss P (2001) Estrogen and the risk of breast cancer. N Engl J Med 344:276–285 [DOI] [PubMed] [Google Scholar]
- Collaborative Group on Hormonal Factors in Breast Cancer (1996) Breast cancer and hormonal contraceptives: collaborative reanalysis of individual data on 53297 women with breast cancer and 100239 women without breast cancer from 54 epidemiological studies. Lancet 347:1713–1727 [DOI] [PubMed] [Google Scholar]
- ——— (1997) Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52705 women with breast cancer and 108411 women without breast cancer. Lancet 350:1047–1059 [PubMed] [Google Scholar]
- Concato J, Feinstein AR, Holford TR (1993) The risk of determining risk with multivariable models. Ann Intern Med 118:201–210 [DOI] [PubMed] [Google Scholar]
- Dupont WD, Page DL, Rogers LW, Parl FF (1989) Influence of exogenous estrogens, proliferative breast disease, and other variables on breast cancer risk. Cancer 63:948–957 [DOI] [PubMed] [Google Scholar]
- Frankel WN, Schork NJ (1996) Who’s afraid of epistasis? Nat Genet 14:371–373 [DOI] [PubMed] [Google Scholar]
- Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF (2000) Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res 60:3440–3444 [PubMed] [Google Scholar]
- Harris JR, Lippman ME, Veronesi U, Willett W (1992) Breast cancer. N Engl J Med 327:319–328 [DOI] [PubMed] [Google Scholar]
- Hayashi S, Watanabe J, Nakachi K, Kawajiri K (1991) Genetic linkage of lung cancer-associated MspI polymorphisms with amino acid replacement in the heme binding region of the human cytochrome P450IA1 gene. J Biochem (Tokyo) 110:407–411 [DOI] [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S (2000) Applied logistic regression. John Wiley & Sons, New York [Google Scholar]
- Kelsey JL, Berkowitz GS (1988) Breast cancer epidemiology. Cancer Res 48:5615–5623 [PubMed] [Google Scholar]
- Kelsey JL, Gammon MD, John EM (1993) Reproductive and hormonal risk factors. Epidemiol Rev 15:36–47 [DOI] [PubMed] [Google Scholar]
- Lachman HM, Papolos DF, Saito T, Yu Y, Szumlanski CL, Weinshilboum RM (1996) Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics 6:243–250 [DOI] [PubMed] [Google Scholar]
- Lavigne JA, Helzlsouer KJ, Huang H, Strickland PT, Bell DA, Selmin O, Watson MA, et al (1997) An association between the allele coding for a low activity variant of catechol-O-methyltransferase and the risk for breast cancer. Cancer Res 57:5493–5497 [PubMed] [Google Scholar]
- Li W, Reich J (2000) A complete enumeration and classification of two-locus disease models. Hum Hered 50:334–349 [DOI] [PubMed] [Google Scholar]
- MacDonald PC, Edman CD, Hemsell DL, Porter JC, Siiteri PK (1978) Effect of obesity on conversion of plasma androstenedione to estrone in postmenopausal women with and without endometrial cancer. Am J Obstet Gynecol 130:448–455 [DOI] [PubMed] [Google Scholar]
- Millikan RC, Pittman GS, Tse CKJ, Duell E, Newman B, Savitz D, Moorman PG, et al (1998) Catechol-O-methyltransferase and breast cancer risk. Carcinogenesis 19:1943–1947 [DOI] [PubMed] [Google Scholar]
- Moore JW, Key TJ, Bulbrook RD, Clark GM, Allen DS, Wang DY, Pike MC (1987) Sex hormone binding globulin and risk factors for breast cancer in a population of normal women who had never used exogenous sex hormones. Br J Cancer 56:661–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M, Kardia SLR, Ferrell RE, Sing CF (2001) A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res 11:458–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parl FF (2000) Estrogens, estrogen receptor and breast cancer. IOS Press, Amsterdam [Google Scholar]
- Peduzzi P, Concato J, Kemper E, Holford TR, Feinstein AR (1996) A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol 49:1373–1379 [DOI] [PubMed] [Google Scholar]
- Persson I, Johansson I, Ingelman-Sundberg M (1997) In vitro kinetics of two human CYP1A1 variant enzymes suggested to be associated with interindividual differences in cancer susceptibility. Biochem Biophys Res Commun 231:227–230 [DOI] [PubMed] [Google Scholar]
- Potischman N, Swanson CA, Siiteri P, Hoover RN (1996) Reversal of relation between body mass and endogenous estrogen concentrations with menopausal status. J Natl Cancer Inst 88:756–758 [DOI] [PubMed] [Google Scholar]
- Rebbeck T, Resvold EA, Duggan DJ, Zhang J, Buetow KH (1994) Genetics of CYP1A1: coamplification of specific alleles by polymerase chain reaction and association with breast cancer. Cancer Epidemiol Biomarkers Prev 3:511–514 [PubMed] [Google Scholar]
- Ripley BD (1996) Pattern recognition and neural networks. Cambridge University Press, Cambridge [Google Scholar]
- Schlichting CD, Pigliucci M (1998) Phenotypic evolution: a reaction norm perspective. Sinauer Associates, Sunderland, MA [Google Scholar]
- Seidegard J, Vorachek WR, Pero RW, Pearson WR (1988) Hereditary differences in the expression of the human glutathione transferase active on trans-stilbene oxide are due to a gene deletion. Proc Natl Acad Sci USA 85:7293–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov I, Akarsu AN, Alozie I, Child A, Barsoum-Hornsy M, Turacli ME, Or M, et al (1998) Sequence analysis and homology modeling suggest primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am J Hum Genet 62:573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen AC, Tilgmann C, Rinne J, Ulmanen I (1997) Genetic polymorphism of catechol-O-methyltransferase (COMT): correlation of genotype with individual variation of S-COMT activity and comparison of the allele frequencies in the normal population and parkinsonian patients in Finland. Pharmacogenetics 7:65–71 [DOI] [PubMed] [Google Scholar]
- Templeton AR (2000) Epistasis and complex traits. In: Wade M, Brodie B III, Wolf J (eds) Epistasis and evolutionary process. Oxford University Press, Oxford, pp 41–57 [Google Scholar]
- Thompson PA, Shields PG, Freudenheim JL, Stone A, Vena JE, Marshall JR, Graham S, et al (1998) Genetic polymorphisms in catechol-O-methyltransferase, menopausal status, and breast cancer risk. Cancer Res 58:2107–2110 [PubMed] [Google Scholar]
- Wade MJ (2000) Epistasis as a genetic constraint within populations and an accelerant of adaptive divergence among them. In: Wade MJ, Brodie B III, Wolf J (eds) Epistasis and evolutionary process. Oxford University Press, Oxford, pp 213–231 [Google Scholar]
- Wiencke JK, Pemble S, Ketterer B, Kelsey KT (1995) Gene deletion of glutathione S-transferase O: correlation with induced genetic damage and potential role in endogenous mutagenesis. Cancer Epidemiol Biomarkers Prev 4:253–259 [PubMed] [Google Scholar]
- Wilson AF, Bailey-Wilson JE, Pugh EW, Sorant AJM (1996) The Genometric Analysis Simulation Program (G.A.S.P.): a software tool for testing and investigating methods in statistical genetics. Am J Hum Genet Suppl 59:A193 [Google Scholar]
- Yager JD, Liehr JG (1996) Molecular mechanisms of estrogen carcinogenesis. Annu Rev Pharmacol Toxicol 36:203–232 [DOI] [PubMed] [Google Scholar]
- Yong LC, Brown CC, Schatzkin A, Schairer C (1996) Prospective study of relative weight and risk of breast cancer: the Breast Cancer Detection Demonstration Project follow-up study, 1979 to 1987-1989. Am J Epidemiol 143:985–995 [DOI] [PubMed] [Google Scholar]