Abstract
Ferritin, which is composed of H and L subunits, plays an important role in iron storage and in the control of intracellular iron distribution. Synthesis of both ferritin subunits is controlled by a common cytosolic protein, iron regulatory protein (IRP), which binds to the iron-responsive element (IRE) in the 5′-UTR of the H- and L-ferritin mRNAs. In the present study, we have identified a single point mutation (A49U) in the IRE motif of H-ferritin mRNA, in four of seven members of a Japanese family affected by dominantly inherited iron overload. Gel-shift mobility assay and Scatchard-plot analysis revealed that a mutated IRE probe had a higher binding affinity to IRP than did the wild-type probe. When mutated H subunit was overexpressed in COS-1 cells, suppression of H-subunit synthesis and of the increment of radiolabeled iron uptake were observed. These data suggest that the A49U mutation in the IRE of H-subunit is responsible for tissue iron deposition and is a novel cause of hereditary iron overload, most likely related to impairment of the ferroxidase activity generated by H subunit.
In white populations, >90% of patients with primary iron overload carry a characteristic mutation, C282Y/H63D, in the hemochromatosis gene (HFE-1) (Feder et al. 1996) or mutation Y250X in the transferrin receptor 2 gene (TFR2, or HFE-3) (Camaschella et al. 2000). However, in Melanesians as well as in Africans and Asians, the gene responsible for primary iron overload is considered to be largely attributable to non-HFE alleles (Monaghan et al. 1998; Sohda et al. 1999).
Ferritin consists of H and L subunits, which assemble to form a shell of 24 subunits with an iron core. Both subunits play important roles in iron storage and in the control of intracellular iron distribution (Theil 1987; Santambrogio et al. 1992; Harrison and Arosio 1996), and it has been shown that each has a distinct role in cellular iron metabolism: H subunit generates ferroxidase activity that appears to be essential for incorporation of iron into the protein shell, whereas L subunit facilitates iron-core formation (Harrison and Arosio 1996). Although H subunit and L subunit are encoded by different genes, located on 11q13 and 19q13.1, respectively (McGill et al. 1987), synthesis of both subunits is controlled by a common cytosolic protein, iron regulatory protein (IRP), which binds to the iron-responsive element (IRE) in the 5′-UTR of the H- and L-subunit mRNAs (Hentze et al. 1987; Leibold and Munro 1988; Thomson et al. 1999; Eisenstein 2000; Theil 2000). In the present study, we demonstrate a single point mutation in the IRE region of H-subunit mRNA, in members of a Japanese family affected with dominantly inherited iron overload.
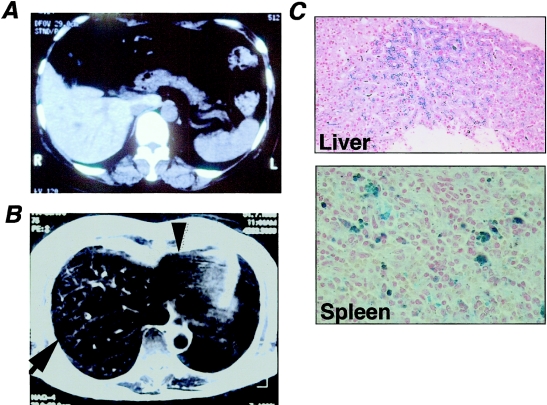
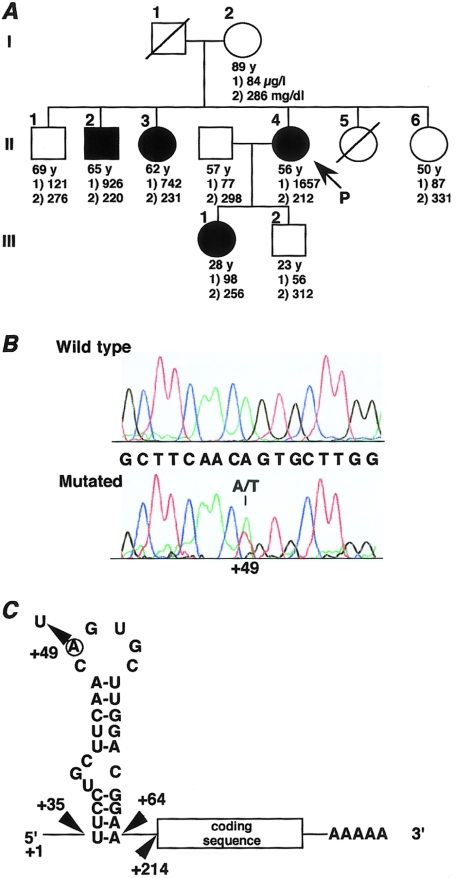
The proband, a woman aged 56 years, was admitted to our hospital for the examination of early gastric cancer that was found, by chance, during a medical examination. An abdominal computed tomography (CT) scan disclosed that the hepatic CT number had increased to 92 units (fig. 1A), and T2-weighted magnetic resonance imaging (MRI) showed low intensity signal of the liver, heart, and bone marrow (fig. 1B). Blood examination showed a high serum ferritin level (1,654 μg/liter) as well as an increase in both serum iron (156 μg/dl) and transferrin saturation (58%) (total iron binding capacity, 269 μg/dl). Hematological examination, including erythrocyte indices, revealed no abnormalities (red blood cell count 4.3 × 1012/liter, hemoglobin 13.3 g/dl, and hematocrit 39%). Blood film showed no siderotic granules (Pappenheimer bodies) in mature erythrocytes. In a specimen obtained by needle liver biopsy, Berlin’s staining revealed heavy iron deposition in most hepatocytes and in some Kupffer cells (fig. 1C, top), suggesting hemochromatosis. The iron deposition was maximal in hepatocytes of zones 1 and 2 of Rappaport, with relative sparing of zone 3. Splenectomy was performed according to a standard surgical procedure for gastric cancer, and iron staining of the specimen also showed gross iron deposition in macrophages (fig. 1C, bottom). Because anemia due to chronic disease was ruled out by (1) absence of anemia, (2) normal range of plasma C-reactive protein and increased transferrin saturation, and (3) a lack of history of iron supplementation, blood transfusion, or alcohol abuse, we suspected a disease relevant to primary hemochromatosis. We then studied seven family members across three generations (fig. 2A) and found that three family members (subjects II-2, II-3, and II-4) had elevated serum ferritin levels (range 742–1,657 μg/liter). There was no history of thalassemia, blood transfusion or donation, abnormal dietary iron intake, alcohol abuse, or pathological iron loss, in any of the affected members. Furthermore, hematologic examination revealed no abnormalities. Abdominal MRI examination was performed on the proband's brother (subject II-2, aged 65 years), and both T1- and T2-weighted MRI showed low signal intensity of the liver and bone marrow, indicating iron deposition (results not shown). Therefore, we suspected genetic factors as the primary cause of hemochromatosis in this family and explored this possibility.
Figure 1.
Abdominal CT and MRI, showing histological features of the liver in an affected member of the family. A, CT of liver in which the CT number increased to 92 units. B, Low-intensity signal of the liver (arrow) and heart (triangle), detected by T2-weighted MRI examination. C, Paraffin-embedded liver biopsy specimen (top, original magnification ×100) and spleen specimen (bottom, original magnification ×200) obtained from the proband, stained with Berlin’s blue (iron staining).
Figure 2.
A, Pedigree of family with dominant primary iron overload. Blackened symbols denote individuals showing the A49T genotype. The numbers under the age indicate concentrations, as follows: 1), serum ferritin (μg/l); 2), serum transferrin concentrations (mg/dl; normal range 205–370 mg/dl). The arrow indicates the proband (II-4). B, Sequence electropherograms and antisense sequence of H-subunit IRE, shown for both a normal (Wild-type) and an affected member (II-4) of the family. PCR products were elecrophoresed on 2% agarose gel, were purified with the Qiaquick Gel Extraction Kit (Qiagen), and were used as templates for fluorescence-based DNA sequence analysis. The mutation is an A→T substitution at position 49, present in the heterozygous state. C, Predicted secondary structure of the IRE motif in the 5′ noncoding region of the H-subunit mRNA. The numbering of bases is from the transcription start site of the human ferritin H chain mRNA (Hentze et al. 1986). The mutation affects the second base of the IRE loop (A49U).
First, we examined the genotype associated with genetic hemochromatosis—specifically, the HFE-1 gene mutation (C282Y and H63D) (Feder et al. 1996) and the TFR2 gene mutation (Y250X) (Camaschella et al, 2000)—by PCR and direct sequence analysis using DNA purified from peripheral blood mononuclear cells. However, neither of these genotypes was detected in this family (results not shown). Normal levels of serum copper and ceruloplasmin activity in family members excluded the possibility of a ceruloplasmin gene defect, which has been shown to be associated with systemic hemosiderosis (Yoshida et al. 1995). Normal levels of serum transferrin (212 mg/dl) and an absence of microcytic anemia also excluded atransferrinemia.
Next, we performed sequencing analysis of H- and L-ferritin cDNAs. Total RNA was extracted from the liver biopsy specimen and was reverse transcribed through use of oligo-dT primer. The entire cDNA sequence of the L and H subunits was amplified by PCR through use of specific primers. Primer sequences were obtained from the Entrez Nucleotide Sequence Search Web site: 5′-GTTGTTGCTTATGATGTGTGA-3′ (forward) and 5′-GTCGTGCTTGAGAGTGAGCCTT-3′ (reverse), for L-subunit; and 5′-CCAGACGTTCTTCGCCGAGA-3′ (forward) and 5′-GCTTTCATTATCACTGTCTC-3′ (reverse), for H-subunit. Amplified DNA fragments were purified and subjected to fluorescence-based DNA sequence analysis. Although the sequence of L-subunit mRNA was normal, we found a heterozygous single A→U conversion at position 49 (A49U) in the second residue of the five-base IRE loop sequence (CAGUG) of the H-subunit mRNA (fig. 2B and C) in four of the family members, suggesting that this mutation was inherited in an autosomal dominant manner (fig. 2A). To search for an A49T mutation in the genomic DNA, we conducted PCR-RFLP analysis using a MunI digestion site that recognizes a mutant IRE sequence (CAATTG) at position 49. However, no A49T mutation was detected in the genomic DNA of 42 unrelated normal subjects. Three of four family members with the A49T genotype had elevated serum ferritin levels. Despite having the A49T genotype, the proband's relatively young daughter (III-1, aged 28 years) showed apparently normal serum ferritin. However, since she had just given birth and was breast feeding at the time of our examination, she was likely to exhibit an increased level of physiological iron loss. In our study, analysis of her case of this inherited disorder leads to the conclusion that iron accumulation may also depend on factors such as iron intake, iron loss, and/or age.
Since IRP has been shown to interact with IRE (Haile et al. 1989; Jaffrey et al. 1993), we performed a competitive binding assay by incubation of IRP from K562 cells with either a [32P]-labeled wild-type IRE probe in the presence of cold wild-type IRE or an A49U-mutated IRE probe. Labeled RNAs were transcribed, in vitro, from an oligonucleotide template, by use of T7 RNA polymerase. The oligonucleotide used to synthesize both the normal IRE probe and the A49U mutated IRE probe for H subunit was as follows: 5′-TTCCGTCCAAGCACTGTTGAAGCAGGAAATCGCCCTATAGTGAGTCGTATTA-3′. The sequence of the oligonucleotide used to synthesize the mutated IRE probe differed by a substitution of A for T in position 49. The underlined sequence represents the T7 RNA polymerase promoter and was double stranded in the transcription reaction, with an oligonucleotide complementary to this region. The RNA used as a probe was labeled with [32P]-UTP to a specific activity of ∼106 cpm/ng. The IRE-[32P]-IRP complex was analyzed by electrophoresis on nondenaturing polyacrylamide gels. The intensity of bands for the IRP-IRE complex diminished more noticeably in the presence of mutated IRE probe than in the presence of wild-type IRE probe (fig. 3A). To evaluate binding affinity between IRE and IRP, we performed direct binding studies using a [32P]-labeled wild-type IRE probe and a [32P]-labeled A49U mutated IRE probe. A Scatchard plot constructed on the basis of these results demonstrated that the mutated IRE showed higher binding affinity to IRP (Kd=13.6 pM) than did the wild-type IRE (Kd=30 pM), suggesting that the mutated IRE binds to IRP rather strongly and thereby causes suppression of H-subunit mRNA translation (fig. 3B).
Figure 3.
Band-shift assay and Scatchard-plot analysis for IRP/IRE interaction. A, Band-shift assay for IRP and either wild-type or mutated IRE. A [32P]-labeled wild-type IRE probe was incubated with 5 μg K562 cytoplasmic extract in the presence of either a cold wild-type IRE probe (lanes 2–4) or a cold mutated IRE probe (lanes 5–7), as a competitor. The IRE-IRP complexes were detected by electrophoresis on nondenaturing 6% polyacrylamide gels and by autoradiography. B, Scatchard-plot analysis for IRP/IRE interaction. Five micrograms of the cytoplasmic extract of K562 cells were incubated with either a [32P]-labeled wild-type IRE probe or a [32P]-labeled mutated probe, in the presence of heparin. Quantification of bound and free RNA was performed by autoradiography on Kodak X-AR film and a densitometry Fast Scan Personal Scanning Imager (Molecular Dynamics). Data were analyzed by the LIGAND program package and fitted to one- or two-binding-site models. The dissociation constant (i.e., Kd) for the high-affinity binding site was calculated.
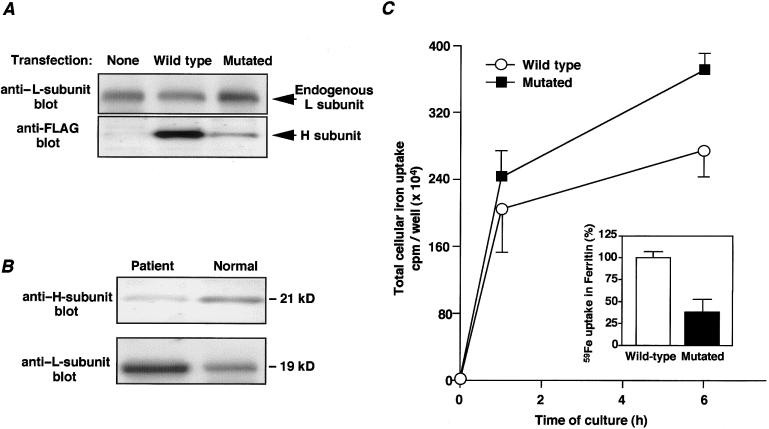
We then constructed two types of FLAG-tagged H-subunit expression vectors, one with mutated IRE and another with wild-type IRE, to examine the effect of the mutated H-subunit expression on the endogenous L-subunit synthesis. C-terminal FLAG-tagged H-subunit expression vector (pFHFr-wt) was constructed by PCR-based subcloning of fragments into pFLAG-CMV-5a. The A49T mutated H-subunit expression vector (pFHFr-mut) was constructed by the cassette mutagenesis method, as described elsewhere (Kawanishi et al. 1995). COS-1 cells were transfected with 1 μg plasmid DNA in the presence of lipofectamine, were lysed 48 h after transfection, and were subjected to immunoblotting with murine anti–L-subunit polyclonal antibodies (M3) or anti–H-subunit monoclonal antibody (HS-59) (Ruggeri et al. 1992). Immunoblots revealed that, in the mutated H-subunit–transfected COS-1 cells, expression of H subunit was suppressed and that of endogenous L subunit was increased in comparison to the wild-type H-subunit transfectant (fig. 4A). When we conducted subunit-specific immunoblotting analysis for liver ferritin, using a patient's (II-4) liver biopsy specimen, expression of H subunit decreased while that of L subunit increased, as compared with expression in normal liver (fig. 4B), consistent with the results of COS-1 transfectants. These results suggest that the A49U mutation of H subunit would affect not only its own translation but also that of L subunit. Since ferritin IREs are known to bind to IRP more effectively than do mRNA IREs such as erythroid 5-aminolevulinic acid and transferrin receptor (Henderson et al. 1994), the increase in L subunit may be due to the preferential uptake of IRP by mutated H-subunit mRNA, allowing high translation efficacy of L subunit.
Figure 4.
A, Effect of mutated H-subunit transfection on expression of endogenous L subunit. A 1-μg plasmid of FLAG-tagged H-subunit expression vector (pFHFr-wt or pFHFr-mut) was transfected to COS-1 cells by use of lipofectamine (Gibco-BRL). Cells were lysed in radioimmunoprecipitation (RIPA) buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) 48 h after transfection, and protein concentration was determined by bicinchoninic-acid assay (Pierce). Ten micrograms of the extract was separated by 15%–25% SDS-PAGE, were transferred to Immobilon-P membranes (Millipore), and were probed with murine anti–L-subunit polyclonal antibodies (M3) or anti–H-subunit monoclonal antibody (HS-59). Antibody-binding sites were detected by the ECL system (Amersham), using horseradish-peroxidase–conjugated goat anti-mouse Ig. B, Subunit-specific immunoblot analysis of liver ferritin in an affected family member (II-4). A portion of the liver specimen was homogenized in RIPA buffer. After centrifugation, 10 μg of the extract from either the patient's liver or a normal liver was subjected to 15%–25% SDS-PAGE and to immunoblotting analysis using anti–H/L-subunit antibodies. C, Effect of mutated H-subunit expression on cellular iron uptake in COS-1 cells. COS-1 transfectants were incubated with 50 μmol/liter [59Fe]Cl3, for 6 h. Cells were lysed with 1% Triton X-100, 10 mmol Tris-HCl/liter, 150 mmol NaCl/liter, pH 7.4. The cell extract was clarified by centrifugation at 1,000 g for 5 min, to remove debris. Cytosolic ferritin was then immunoprecipitated using anti-FLAG antibody-immobilized agarose beads. Total cellular 59Fe uptake and 59Fe incorporation into ferritin molecules were determined by gamma counting. Error bars represent the mean ± SD of three determinations.
The CAGUG loop structure in the IRE is well conserved in all ferritin mRNAs examined in vertebrates (Aziz and Munro 1987). In vitro studies using synthetic IRE-analogue oligonucleotides have demonstrated that the CAGUG sequence in the loop, as well as base pairing along the upper stem, are crucial for IRE/IRP interaction (Jaffrey et al. 1993; Henderson et al. 1994), and substitution or deletion of a single base of IRE (especially in the loop sequence) results in a substantial decrease in binding affinity (6–654-fold) (Jaffrey et al. 1993). These results from in vitro data are clinically supported by recent studies of hereditary hyperferritinemia-cataract syndrome (HHCS), in which point mutations in the loop or bulge of the IRE of L-subunit mRNA resulted in a loss of IRE affinity to IRP and in enhanced translation of L subunit, neither of which were related to iron overload (Beaumont et al. 1995; Cazzola et al. 1997; Levi et al. 1998). Our study is the first demonstration of an increment in IRE affinity to IRP caused by a point mutation at position 49 of H-subunit mRNA.
With regard to tissue iron deposition in affected individuals, we postulated that the decrease in H subunit might be responsible, since H subunit is known to generate a ferroxidase activity that is essential for the incorporation of iron into the ferritin molecule (Harrison and Arosio 1996). To test this assumption, we performed an iron-uptake assay using the aforementioned COS-1 transfectants. After transient expression of the mutated H subunit or wild-type H subunit in COS-1 cells, [59Fe]Cl3 was added to the culture medium. The cells were solubilized, and cytosolic ferritin was immunoprecipitated through use of anti-FLAG antibody. In the mutated H-ferritin transfectant, 59Fe incorporation into the ferritin molecule was significantly lower and total cellular uptake of iron was higher than they were in the wild-type H-ferritin transfectant (fig. 4C). This result suggested that the decrease in H subunit not only impaired iron uptake into the ferritin molecule but also caused iron accumulation in the cytosol, possibly because of the decrease in ferroxidase activity. This assumption has been verified by a recent study (Ferreira et al. 2000), in which H-ferritin gene–knockout mice showed early embryonic lethality due to massive iron deposition. In this respect, it is of interest that no iron overload is associated with HHCS (Beaumont et al. 1995), despite L subunit being similarly increased; this is probably because of the fact that H subunit is not affected. Furthermore, the effect of the mutation in the L-subunit IRE in patients with HHSC is associated with a relatively benign phenotype: early onset of cataracts (Beaumont et al. 1995).
In the present study, we have characterized an A49T mutation in the IRE sequence of the H-ferritin gene as a novel, inherited genetic abnormality related to iron overload. Since the H-ferritin gene and genes for IRE-binding proteins have been reported to be unrelated to chromosome 6p (the locus for HFE hemochromatosis) (Zheng et al. 1994), this genotype may be associated with non-HFE-type iron overload. In this regard, it may be noteworthy that autosomal dominant and non-HFE-type iron overload has been reported elsewhere, in three Japanese families (Kawanaka et al. 1998). Additional family studies are certainly needed to elucidate whether this mutation is an isolated or a common one.
Acknowledgments
We thank Ms. Sachie Miyake, for technical assistance; Dr. Paolo Arosio, for providing anti-ferritin antibodies; and Kevin Litton, B.A., for editorial assistance.
Electronic-Database Information
The accession numbers and URLs for data in this article are as follows:
- Entrez-Nucleotide, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide (for the H-ferritin cDNA sequence [accession number L20941]; the L-ferritin cDNA sequence [accession number M10119]; and the genomic sequence for the HFE gene [accession number U60319])
References
- Aziz N, Munro HN (1987) Iron regulates ferritin mRNA translation through a segment of its 5′ untranslated region. Proc Natl Acad Sci USA 84:8478–8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont C, Leneuve P, Devaux I, Scoazec JY, Berthier M, Loiseau MN, Grandchamp B, Bonneau D (1995) Mutation in the iron responsive element of the L ferritin mRNA in a family with dominant hyperferritinaemia and cataract. Nat Genet 11:444–446 [DOI] [PubMed] [Google Scholar]
- Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P (2000) The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 25:14–15 [DOI] [PubMed] [Google Scholar]
- Cazzola M, Bergamaschi G, Tonon L, Arbustini E, Grasso M, Vercesi E, Barosi G, Bianchi PE, Cairo G, Arosio P (1997) Hereditary hyperferritinemia-catracta syndrome: Relationship between phenotypes and specific mutations in the iron-responsive element of ferritin light-chain mRNA. Blood 90:814–821 [PubMed] [Google Scholar]
- Eisenstein RS (2000) Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr 20:627–662 [DOI] [PubMed] [Google Scholar]
- Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, et al (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13:399–408 [DOI] [PubMed] [Google Scholar]
- Ferreira C, Bucchini D, Martin ME, Levi S, Arosio P, Grandchamp B, Beaumont C (2000) Early embryonic lethality of H ferritin gene deletion in mice. J Biol Chem 275:3021–3023 [DOI] [PubMed] [Google Scholar]
- Haile DJ, Hentze MW, Rouault TA, Harford JB, Klausner RD (1989) Regulation of interaction of the iron-responsive element binding protein with iron-responsive RNA elements. Mol Cell Biol 9:5055–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P (1996) The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta 1275:161–203 [DOI] [PubMed] [Google Scholar]
- Henderson BR, Menotti E, Bonnard C, Kühn LC (1994) Optimal sequence and structure of iron-responsive elements. J Biol Chem 269:17481–17489 [PubMed] [Google Scholar]
- Hentze MW, Caughman SW, Rouault TA, Barriocanal JG, Dancis A, Harford JB, Klausner RD (1987) Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science 238:1570–1573 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Keim S, Papadopoulos P, O'Brien S, Modi W, Drysdale J, Leonard WJ, Harford JB, Klausner RD (1986) Cloning, characterization, expression, and chromosomal localization of a human ferritin heavy-chain gene. Proc Natl Acad Sci USA 83:7726–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Haile DJ, Klausner RD, Harford JB (1993) The interaction between the iron-responsive element binding protein and its cognate RNA is highly dependent upon both RNA sequence and structure. Nucleic Acids Res 21:4627–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanaka K, Kinoyama S, Niiyama G, Kimura T, Ifukube S, Onogi T, Miyake I, Tamura T, Mitani K, Sugahara A, Goto K, Uchida J, Ito T, Yamada G, Moriya T (1998) A case of idiopathic hemochromatosis which occurred in three siblings with high level of serum CA 19-9. Nippon Shokakibyo Gakkai Zasshi 95:910–915. [PubMed] [Google Scholar]
- Kawanishi J, Kato J, Sasaki K, Fujii S, Watanabe N, Niitsu Y (1995) Loss of E-cadherin-dependent cell-cell adhesion due to mutation of the β-catenin gene in a human cancer cell line, HSC-39. Mol Cell Biol 15:1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold EA, Munro HN (1988) Cytoplasmic protein binds to in vitro to a highly conserved sequence in the 5′ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci USA 85:2171–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S, Girelli D, Perrone F, Pasti M, Beaumont C, Corrocher R, Albertini A, Arosio P (1998) Analysis of ferritins in lymphoblastoid cell lines and in the lens of subjects with hereditary hyperferritinemia-catract syndrome. Blood 91:4180–4187 [PubMed] [Google Scholar]
- McGill JR, Naylor SL, Sakaguchi AY, Moore CM, Boyd D, Barrett KJ, Shows TB, Drysdale JW (1987) Human ferritin H and L sequences lie on ten different chromosomes. Hum Genet 76:66–72 [DOI] [PubMed] [Google Scholar]
- Monaghan KG, Rybicki BA, Shurafa M, Feldman GL (1998) Mutation analysis of the HFE gene associated with hereditary hemochromatosis. Am J Hematol 58:213–217 [DOI] [PubMed] [Google Scholar]
- Ruggeri G, Santambrogio P, Bonfiglio F, Levi S, Bugari G, Verardi R, Cazzola M, Invernizzi R, Zambelli LM, Albertini A, Arosio P (1992) Antibodies for denatured human H-ferritin stain only reticuloendothelial cells within the bone marrow. Br J Haematol 81:118–124 [DOI] [PubMed] [Google Scholar]
- Santambrogio P, Levi S, Arosio P, Palagi L, Vecchio G, Lawson DM, Yewdall SJ, et al (1992) Evidence that a salt bridge in the light chain contributes to the physical stability difference between heavy and light human ferritins. J Biol Chem 267:14077–14083 [PubMed] [Google Scholar]
- Sohda T, Yanai J, Soejima H, Tamura K (1999) Frequencies in the Japanese population of HFE gene mutations. Biochem Genet 37:63–68 [DOI] [PubMed] [Google Scholar]
- Theil EC (1987) Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem 56:289–315 [DOI] [PubMed] [Google Scholar]
- ——— (2000) Targeting mRNA to regulate iron and oxygen metabolism. Biochem Pharmacol 59:87–93 [DOI] [PubMed] [Google Scholar]
- Thomson AM, Rogers JT, Leedman PJ (1999) Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int J Biochem Cell Biol 31:1139–1152 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Furihata K, Takeda S, Nakamura A, Yamamoto K, Morita H, Hiyamuta S, Ikeda S, Shimizu N, Yanagisawa N (1995) A mutation in the ceruloplasmin gene is associated with systemic hemosiderosis in humans. Nat Genet 9:267–272 [DOI] [PubMed] [Google Scholar]
- Zheng H, Bhavsar D, Volz A, Ziegler A, Drysdale J (1994) Exclusion of ferritins and iron-responsive element (IRE)-binding proteins as candidates for hemochromatosis gene. Hum Genet 94:159–164 [DOI] [PubMed] [Google Scholar]