Abstract
Otopalatodigital syndrome type 1 (OPD1) is an X-linked semidominant condition characterized by malformations of the skeleton, auditory apparatus, and palate. Previous studies have established linkage to a 16-cM region of Xq27-q28. A proposed allelic variant of OPD1, termed “OPD2,” is associated with a more severe, frequently lethal phenotype with visceral and brain anomalies in addition to skeletal, auditory, and palatal defects. We report linkage of the OPD2 phenotype to a 2-cM region of distal Xq28 in a Maori kindred, with a maximum multipoint LOD score of 3.31 between the markers DXS1073 and DXS1108. This provides support for allelism between OPD1 and OPD2 and reduces the size of the disease interval to 1.8–2.1 Mb. We also demonstrate that female carriers of this disorder exhibit skewed inactivation that segregates with the high-risk haplotype and may be inversely related to the severity with which they manifest features of the disorder.
Otopalatodigital syndrome type 1 (OPD1 [MIM 311300]) is an X-linked condition characterized, in affected males, by skeletal abnormalities, conductive or sensorineural hearing loss, and cleft palate (Taybi et al. 1962; Dudding et al. 1967). The skeletal dysplasia comprises camptodactyly, long spatulate fingers, pectus carinatum, mild campomelia, and malformed auditory ossicles—the latter feature leading to conductive hearing loss in some individuals. Carrier females may exhibit the same manifestations as males but generally have a milder range of expressivity. A second entity—comprising a more severe skeletal dysplasia, cleft palate, and micrognathia—was subsequently described (Fitch et al. 1976) and was later designated “otopalatodigital syndrome type 2” (OPD2 [MIM 304120]), owing to its clinical similarity to OPD1 (Fitch et al. 1983). Subsequent reports have extended the phenotype of OPD2 to include cardiac, genitourinary, gastrointestinal, and central nervous system anomalies. Perinatal death is common because of these malformations, and intellectual disability sometimes occurs, in contrast to OPD1. As in OPD1, carrier females with OPD2 exhibit a phenotype that can extend from a mild subclinical osteodysplasia to a presentation indistinguishable from that of affected males.
Two studies have previously linked OPD1 to the distal portion of the long arm of the X chromosome, between DXS539 (located in Xq27) and the telomere—a distance of 16 cM (Biancalana et al. 1991; Hoar et al. 1992). No linkage studies have previously been performed on OPD2, and, therefore, no experimental evidence exists to support the clinical hypothesis that these two disorders are allelic.
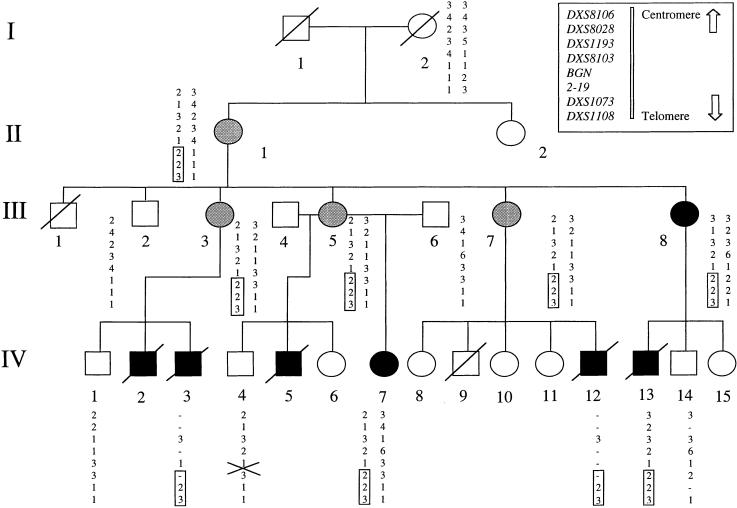
We have previously ascertained a four-generation family of Maori ancestry from New Zealand that segregates OPD2 (Robertson et al. 1997). Several individuals have been born since the original report, and the updated pedigree is presented in figure 1. Within this family, there have been five affected males, all of whom died perinatally and had a severe skeletal dysplasia with campomelia, thoracic constriction, and digital anomalies. Three of these individuals had an omphalocele, two had cleft palate, and four had genitourinary anomalies. An additional male (III-1) died perinatally of unknown cause. Six females could be identified as carriers, either because they had transmitted the mutant allele (four subjects), because they manifested specific features of OPD2 (one subject), or for both reasons (one subject). The specific manifestations were conductive deafness caused by malformed auditory ossicles (III-8) and bilateral hearing loss, cleft palate, spatulate fingers, and a chest wall deformity (IV-7). The four apparently healthy carriers exhibited minor, but consistent, radiological anomalies of the skull base, ribs, vertebral bodies, and long bones. The study was approved by the local institutional ethics committee, and informed consent was given to obtain venous blood, from all available members of the pedigree, and fixed tissue, from autopsy specimens of IV-3 and IV-12. DNA was prepared using standard techniques.
Figure 1.
Pedigree of the family segregating for OPD2. Unblackened circle, unaffected female; gray circle, subclinically affected female; blackened circles, clinically manifesting female; and blackened square, affected male. Haplotypes are depicted for markers located in the distal region of Xq, and the segment that segregates perfectly with the disease is boxed. Individual III-8 is known to have a different father from her female siblings. Genotypes for the locus BGN represent a combination of results obtained from DXSBGN and the novel SNP identified in intron 5. The locus 2–19 is a novel marker identified as polymorphic during the course of this study (see text). The position of the key recombination in individual IV-4 is indicated with a cross. The results for clinically unaffected females in generation IV are omitted, to avoid revealing their carrier status.
Given the established clinical and pedigree evidence that OPD2 segregates as an X-linked trait, the segregation of a panel of polymorphic markers encompassing the entire X chromosome (Dib et al. 1996) was studied in this family. The data were analyzed using the MLINK and LINKMAP programs from the FASTLINK 4.0 software package (Lathrop and Lalouel 1984; Cottingham et al. 1993; Schäffer et al. 1994), available through the U.K. Human Genome Mapping Project, to obtain two-point and multipoint LOD scores, respectively. The trait was assigned as X linked, with the penetrance of the disease allele set at 1.0 and with the disease-allele frequency set at .0001. All males and the six females identified as carriers were included in the calculation of the LOD score.
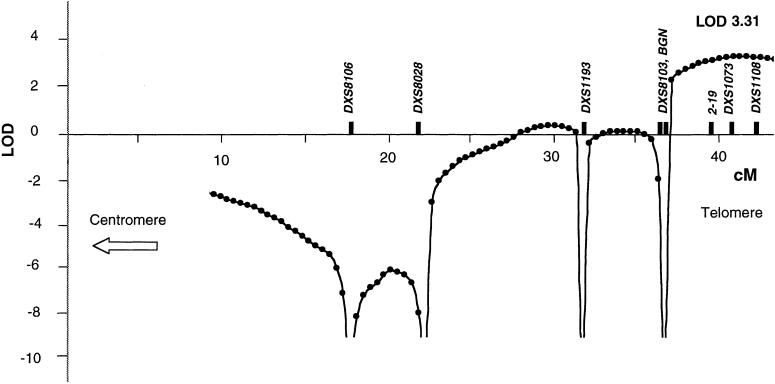
The only markers that yielded significantly positive LOD scores mapped to distal Xq28. Markers DXS1073 and DXS1108, located 1.5 Mb and 0.5 Mb from the telomere, respectively, gave two-point LOD scores of 2.41 and 2.71, respectively, at θ=0. By multipoint analysis, a maximum LOD score of 3.31 was obtained between markers DXS1073 and DXS1108 (fig. 2). Localization to any other region of the X chromosome was excluded, at odds of >100:1 (data not shown). This confirmed that the OPD2 phenotype in this family maps to the same region of the X chromosome as does OPD1, consistent with allelism of the two conditions.
Figure 2.
Multipoint LOD analysis of the family, in the region of the Xq telomere
Inspection of haplotypes identified a recombination at DXS8103 in the unaffected male IV-4, placing the disease locus distal to this marker (fig.1). We evaluated BGN, which maps distal to DXS8103 and encodes the proteoglycan biglycan, as a candidate disease gene (Fisher et al. 1991). However, we identified a previously undescribed single-nucleotide polymorphism (SNP) (IVS5+90C→T), for which IV-4 had inherited the same allele that was found in affected individuals IV-3 and IV-7, excluding BGN as the disease gene and placing the disease gene distal to this locus (fig. 1).
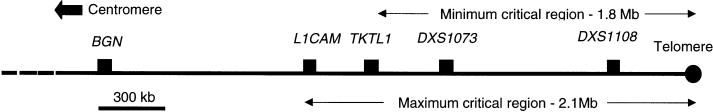
To refine the linkage interval more precisely, we searched published sequences within the region bounded by BGN and DXS1073, for repetitive sequences and SNPs. No published SNP or polymorphism was heterozygous in III-5, but we identified three novel polymorphisms that were informative for the critical meiosis. An (AT)n repeat was positioned 5′ to the gene 2-19, which is located 1.6 Mb from the telomere, and was amplified with the primers 5′-GCAACAAAGTGAGACCCTGC-3′ and 5′-GTTGGCTGTGTCAGGTCAGG-3′ (GenBank L44140). The second locus, a polyadenine stretch located adjacent to the TKTL1 gene 1.8 Mb from the telomere, was amplified with the primers 5′-CGTGGTGGCTTACACCTGTC-3′ and 5′-ATACCTGCTTTCGTCGGGGCACAC-3′ (GenBank Z49258). Both these loci were shown to be nonrecombinant in the unaffected male (IV-4). The disease allele at locus 2-19 originated from the unaffected great-grandfather, I-1, indicating that the mutation arose de novo in patient II-1 (fig. 1). A locus comprising a complex CnTn repeat stretching over ∼350 bp, lying 1.5 kb 3′ to the L1CAM gene (GenBank U52112), was also identified as polymorphic. This repeat was amplified initially using the primers 5′-AAGTTCTCACCTTGAAAGTGCAG-3′ and 5′-GTAAAAAAATCAGGTTGCAGCG-3′ and was followed by a second reaction employing primers 5′-AAGTGGAGGGCTCACCTGTG-3′ and 5′-CGACAGAGCGAGACAAAGAAAG-3′. The two alleles in patient III-5 differed in length by a single CT dinucleotide within the repeat, and individual IV-4 was recombinant at this locus. This analysis reduced the OPD2 interval to a maximum of 2.1 Mb (L1CAM–Xqter) and a minimum of 1.8 Mb (TKTL1–Xqter) (fig. 3).
Figure 3.
Physical map outlining the position and extent of the candidate interval containing the gene mutated in OPD2
The terminal 2.1 Mb of Xq has been intensively studied, and much of it is sequenced and characterized in terms of its gene content. Mutations in 12 genes are known to cause specific monogenic disorders, and many additional diseases map to this general region. The density of genes within the candidate interval is uneven; gene-dense segments are found within the proximal 500 kb, but more-distal sequences have been predicted to be relatively gene poor (De Sario et al. 1996). The 400 kb bounded by the telomere, termed the “Xq/Yq homology region,” is fully sequenced and encodes four genes and one pseudogene (Ciccodicola et al. 2000). Although all four genes have counterparts on the homologous portion of the Y chromosome, some are inactivated in a Y-specific manner. This, together with the substantially lower recombination rate compared to that observed for the primary pseudoautosomal region at Xp/Yp, means that these genes remain potential candidates for a disorder such as OPD.
The linkage of OPD2 to a narrow region within the candidate interval previously identified for OPD1 provides the first experimental support for the hypothesis that the two conditions are allelic. Clinical evidence exists to suggest that two other conditions, frontometaphyseal dysplasia (FMD [MIM 305620]) and Melnick-Needles syndrome (MNS [MIM 309350]) may also be allelic to OPD types 1 and 2 (Verloes et al. 2000). FMD segregates as an X-linked semidominant condition and is characterized by skull hyperostosis, digital/skeletal anomalies, and deafness and is therefore reminiscent of OPD1 (Superti-Furga and Gimelli 1987). Affected males born to women with MNS bear striking resemblance to males with OPD2 (Verloes et al. 2000). It is therefore possible that the 1.8–2.1-Mb region that we have identified as the candidate interval for OPD2 contains the gene mutated in all four conditions.
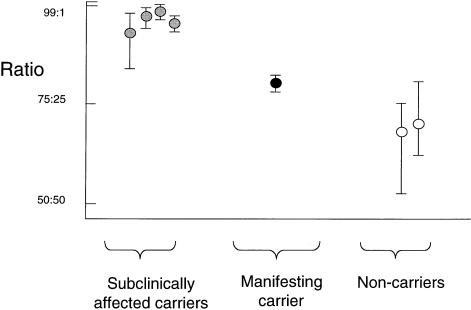
To gain further insight into the expression of the OPD2 disease phenotype in carrier females, we performed X-inactivation studies on DNA extracted from peripheral blood samples. The relative methylation (and, hence, inactivation) of the two alleles of the androgen-receptor CAG repeat was measured by quantifying each allele after digestion of the radiolabeled PCR product by the restriction enzyme HpaII (Allen et al. 1992). Band intensity was captured on a phosphoimager screen and was integrated using IMAGEQUANT software, version 5.1 (Molecular Dynamics). Two of the four obligate carriers with only subclinical evidence of their carrier status were informative in this assay and demonstrated >90% inactivation of one allele in blood leucocytes (fig. 4). Individual IV-7, who has a cleft palate, deafness, and moderately severe skeletal findings, was less skewed (80% inactivation of one allele). Individual III-8 with conductive deafness caused by ossicular deformities was uninformative. Two other females in generation IV, who are clinically normal but have not been examined radiologically, are predicted, by linkage analysis, to carry the mutant allele and were also noted to be >90% skewed. Two females (I-2 and another in generation IV), predicted, by linkage analysis, to be noncarriers demonstrated a random X-inactivation pattern (68% and 71%, respectively).
Figure 4.
Skewing of X inactivation (relative methylation of alleles at the androgen-receptor locus) in relation to affection status of females in the pedigree. Each symbol marks the mean of three independent experiments, with the range of values obtained shown by a vertical bar. Definitions of symbols are the same as in figure 1, except that two individuals in generation IV, predicted by linkage analysis to be carriers, are represented by gray symbols.
The preponderance of severely skewed individuals in this family is well in excess of the proportion observed in the general population (Sharp et al. 2000). Moreover, skewed X inactivation cosegregates with the OPD2 and high-risk haplotype. The most likely explanation for these observations is that, in carrier females, selection occurs early in embryogenesis, against cells in which the active X chromosome bears the mutant allele, as is documented in several other X-linked disorders (Migeon and Haisley-Royster 1998).
The manifesting female IV-7 showed a less dramatically skewed pattern of X inactivation. Variation in the severity of the female phenotype has been described for both OPD1 (Gall et al. 1972; Gorlin et al. 1973) and OPD2 (Robertson et al. 1997), and these data suggest that variation in the degree of X inactivation may be responsible for these observations, as has been proposed for the X-linked dominant condition Rett syndrome (Amir et al. 2000). Once the causative gene is identified, study of a larger group of unrelated male and female patients with OPD and of its allelic conditions will allow description not only of genotypic but also of epigenetic influences, such as X inactivation, on the phenotypic spectrum observed in these syndromes.
Acknowledgments
We thank the members of the family, for their support for this study; Drs. I. Winship and M. Delatycki, for patient samples; and Dr. S. Twigg, for technical support. Computing facilities for linkage analysis were provided by the U.K. Human Genome Mapping Project Resource Centre. The work was funded by a Nuffield Medical Fellowship (S.P.R.) and the Wellcome Trust (A.O.M.W.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for primers ajacent to the L1CAM, 2-19, and TKTL1 genes)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OPD1 [MIM 311300], OPD2 [MIM 304120], FMD [MIM 305620], and MNS [MIM 309350])
References
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239 [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Schultz R, Malicki DM, Tran CQ, Dahle EJ, Philippi A, Timar L, Percy AK, Motil KJ, Lichtarge O, Smith EO, Glaze DG, Zoghbi HY (2000) Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann Neurol 47:670–679 [PubMed] [Google Scholar]
- Biancalana V, le Marec B, Odent S, van den Hurk JA, Hanauer A (1991) Oto-palato-digital syndrome type1: further evidence for assignment of the locus to Xq28. Hum Genet 88:228–230 [DOI] [PubMed] [Google Scholar]
- Ciccodicola A, D'Esposito M, Esposito T, Gianfrancesco F, Migliaccio C, Miano MG, Matarazzo MR, Vacca M, Franzè A, Cuccurese M, Cocchia M, Curci A, Terracciano A, Torino A, Cocchia S, Mercadante G, Pannone E, Archidiacono N, Rocchi M, Schlessinger D, D'Urso M (2000) Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet 9:395–401 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- De Sario A, Geigl E-M, Palmieri G, D’Urso M, Bernardi G (1996) A compositional map of human chromosome band Xq28. Proc Natl Acad Sci USA 93:1298–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dudding BA, Gorlin RJ, Langer LO (1967) The oto-palato-digital syndrome: a new symptom complex consisting of deafness, dwarfism, cleft palate, characteristic facies, and a generalized bone dysplasia. Am J Dis Child 113:214–221 [DOI] [PubMed] [Google Scholar]
- Fisher LW, Heegaard A-M, Vetter U, Vogel W, Just W, Termine JD, Young MF (1991) Human biglycan gene: putative promoter, intron-exon junctions, and chromosomal localization. J Biol Chem 266:14371–14377 [PubMed] [Google Scholar]
- Fitch N, Jequier S, Gorlin R (1983) The otopalatodigital syndrome, proposed type II. Am J Med Genet 15:655–664 [DOI] [PubMed] [Google Scholar]
- Fitch N, Jequier S, Papageorgiou A (1976) A familial syndrome of cranial, facial, oral and limb anomalies. Clin Genet 10:226–231 [DOI] [PubMed] [Google Scholar]
- Gall JC, Stern AM, Poznanski AK, Garn SM, Weinstein ED, Hayward JR (1972) Oto-palato-digital syndrome: comparison of clinical and radiographic manifestations in males and females. Am J Hum Genet 24:24–36 [PMC free article] [PubMed] [Google Scholar]
- Gorlin R, Poznanski A, Hendon I (1973) The oto-palato-digital (OPD) syndrome in females. Oral Surg Oral Med Oral Pathol 35:218–224 [DOI] [PubMed] [Google Scholar]
- Hoar DI, Field LL, Beards F, Hoganson G, Rollnick B, Hoo JJ (1992) Tentative assignment of gene for otopalatodigital syndrome to distal Xq(Xq26-28). Am J Med Genet 42:170–172 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculation of lod scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Haisley-Royster C (1998) Familial skewed X inactivation and X-linked mutations: unbalanced X inactivation is a powerful means to ascertain X-linked genes that affect cell proliferation. Am J Hum Genet 62:1555–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S, Gunn T, Allen B, Chapman C, Becroft D (1997) Are Melnick-Needles syndrome and otopalatodigital syndrome type II allelic? observations in a four generation kindred. Am J Med Genet 71:341–347 [DOI] [PubMed] [Google Scholar]
- Schäffer AA, Gupta SK, Shriram K, Cottingham RW (1994) Avoiding recomputation in linkage analysis. Hum Hered 44:225–237 [DOI] [PubMed] [Google Scholar]
- Sharp A, Robinson D, Jacobs P (2000) Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet 107:343–349 [DOI] [PubMed] [Google Scholar]
- Superti-Furga A, Gimelli F (1987) Fronto-metaphyseal dysplasia and the oto-palato-digital syndrome. Dysmorph Clin Genet 1:2–5 [Google Scholar]
- Taybi H (1962) Generalized skeletal dysplasia with multiple anomalies: a note on Pyle’s disease. Am J Roentgen 88:450–457 [PubMed] [Google Scholar]
- Verloes A, Lesenfants S, Barr M, Grange DK, Journel H, Lombet J, Mortier G, Roeder E (2000) Fronto-otopalatodigital osteodysplasia: clinical evidence for a single entity encompassing Melnick-Needles syndrome, otopalatodigital syndrome types 1 and 2, and frontometaphyseal dysplasia. Am J Med Genet 90:407–422 [DOI] [PubMed] [Google Scholar]