Abstract
Usher syndrome type IIa is an autosomal recessive disorder characterized by mild-to-severe hearing loss and progressive visual loss due to retinitis pigmentosa. The mutation that most commonly causes Usher syndrome type IIa is a 1-bp deletion, described as “2299delG,” in the USH2A gene. The mutation has been identified in several patients from northern and southern Europe and from North America, and it has been found in single patients from South America, South Africa, and China. Various studies have reported a range of frequencies (.16–.44) among patients with Usher syndrome, depending on the geographic origin of the patients. The 2299delG mutation may be the one that most frequently causes retinitis pigmentosa in humans. Given the high frequencies and the wide geographic distribution of the mutation, it was of interest to determine whether the mutation resulted from an ancestral mutational event or represented a mutational hotspot in the USH2A gene. Haplotype analysis was performed on DNA samples from 116 unrelated patients with Usher syndrome type IIa; the patients were from 14 countries and represented 148 2299delG alleles. On the basis of six single-nucleotide polymorphisms within the USH2A gene, 12 core haplotypes were observed in a panel of normal chromosomes. However, in our analysis, only one core haplotype was found to be associated with the 2299delG mutation. The data indicate that the widespread geographic distribution of the 2299delG mutation is the result of an ancestral mutation that has spread throughout Europe and into the New World as a result of migration.
The Usher syndromes (MIM 276900) are a group of recessively inherited disorders characterized by progressive visual loss caused by retinitis pigmentosa (RP) and by varying degrees of hearing impairment (which may occur in the presence or absence of vestibular dysfunction) (Smith et al. 1994). The syndromes constitute the most frequent cause of acquired deaf-blindness, with a prevalence of 4.4:100,000 in the United States (Boughman et al. 1983) and 2.2:100,000 in Denmark (Rosenberg et al. 1997). Usher syndrome has been subcategorized into three distinct clinical types and has been linked to 10 distinct loci, USH1A–1F, USH2A–2C, and USH3 (Hereditary Hearing Loss Home page). Usher type II probably accounts for more than half of all Usher cases. Since the time of the initial identification of mutations in the USH2A gene, which are responsible for Usher syndrome type IIa and nonsyndromic retinitis pigmentosa (RP) (Eudy et al. 1998; Rivolta et al. 2000), a total of 31 distinct disease-causing mutations have been reported (Adato et al. 2000; Dreyer et al. 2000; Weston et al. 2000). Most of the mutations are private. However, the 2299delG mutation, in exon 13, stands out because of its high frequency (.16–.44) across various studies (Beneyto et al. 2000; Dreyer et al. 2000; Liu et al. 1999; Weston et al. 2000). The mutation has been identified in patients from northern and southern Europe and North America and in single patients from South America, South Africa, and China.
Given the high frequencies of the 2299delG mutation among patients with Usher type IIa and the mutation's wide geographic distribution, it was of interest to investigate whether the 2299delG alleles have a common origin or represent a hotspot in the USH2A gene. We therefore analyzed DNA samples obtained from 116 unrelated patients with Usher type II, of whom 32 were homoallelic and 84 were heteroallelic for the 2299delG mutation. The patients originated in 14 countries: the United States (43 patients), Denmark (22 patients), The Netherlands (17 patients), United Kingdom (10 patients), Spain (6 patients), Sweden (6 patients), Norway (4 patients), Belgium (2 patients), Canada (1 patient), China (1 patient), Colombia (1 patient), France (1 patient), Germany (1 patient), and South Africa (1 patient); all the United States patients were of European ancestry, except for one African American.
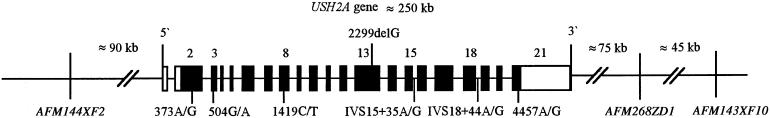
To establish the haplotype for the 2299delG alleles, six single-nucleotide polymorphisms (SNPs) within the USH2A gene, exons 2–21, were selected (Dreyer et al. 2000). Two SNPs (IVS15+35A/G and IVS17−8T/G) were in noncoding sequence. Four SNPs were in coding sequence, of which two were synonymous (504G/A and 1419C/T) and two were nonsynonymous (373A/G and 4457A/G). In addition, three flanking markers, AFM144XF2, AFM268ZD1, and AFM143XF10, which span ∼460 kb of the USH2A locus, were included (fig. 1). The allele frequencies of these SNPs were established in normal chromosomes from Norwegian nuclear families (n=108), Spanish control chromosomes (n=94), and in normal chromosomes from parents of patients with Usher syndrome (n=49). There were no significant differences in allele frequencies among the three control panels (data not shown).
Figure 1.
Relative locations of the SNPs and the 2299delG mutation within the USH2A gene and the three selected flanking markers. Exons 1–21 are depicted as boxes and are numbered intermittently, for the sake of legibility. We used distance estimates that were derived from Weston et al. (2000) and the Genome Database.
SNP analysis was performed by PCR amplification in a standard reaction mixture, as described by Dreyer et al. (2000), except IVS15+35A/G, which was detected by allele-specific PCR using primers (reverse: 5′-GCAGTCCCCTGTATGATGATGC-3′; reverse: 5′-ATGCAGTCCCCTGTATGATGGTGT-3′; and forward: 5′-AAGCCGTCTTACTCTACAATGCT-3′). The products were 321 bp and 323 bp for IVS15+35G and IVS15+35A, respectively. Fluorescence-labeled reverse primers were used, and products were analyzed on a 377-ABI sequencer (Applied Biosystems). The underlined mismatches were introduced to increase the specificity of the PCR. Genotyping for the three markers (AFM144XF2, AFM268ZD1, and AFM143XF10) was performed by use of information from the Genome Database. The determination of haplotype phase was based either on homozygosity or on the analysis of parental samples, which could be used fully or partially to determine the phase. A complete description of the USH2A haplotypes for the 148 USH2A alleles is presented in table 1.
Table 1.
Haplotypes Associated with the 2299delG Alleles in 116 Unrelated Patients of Geographically Diverse Origins[Note]
|
Genotype at |
|||||||||
| Country and Patient Number | AFM144 XF2 | 373A/G | 504G/A | 1419C/T | IVS15+35A/G | IVS17−8T/G | 4457A/G | AFM268ZD1 | AFM143XF10 |
| United States: | |||||||||
| 791a | 116 | A | G | C | A | T | A | 250 | 111 |
| 229 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| 1143 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| 2140 | 116 | A | G | C | A | T | A | 250 | 111 |
| 116 | A | G | C | A | T | A | 250 | 111 | |
| 2720 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| 52 | 116 | A | G | C | A | T | A | 250 | 113 |
| 296 | 118/126 | A | G | C | A | T | A | 250 | 111 |
| 427 | 116 | A | G | C | A | T | A | 250 | 111 |
| 560 | 116/126 | A | G | C | A | T | A | 250 | 111/115 |
| 685 | 126 | A | G | C | A | T | A | 250 | 111 |
| 747 | 116 | A | G | C | A | T | A | 250 | 111 |
| 777 | 116/126 | A | G | C | A | T | A | 250 | 111/115 |
| 795 | 116 | A | G | … | A | T | A | 250 | 111 |
| 869 | 116 | A | G | C | A | T | A | 250 | 119 |
| 876 | 116/126 | A | G | C | A | T | A | 250 | 109/111 |
| 225 | 126 | A | G | C | A | T | A | 250 | 111/113 |
| 927 | 116 | A | G | C | A | T | A | 250 | 119 |
| 1052 | 116/126 | A | G | C | A | T | A | 250 | 107/111 |
| 1121 | 116/126 | A | G | C | A | T | A | 250/254 | 111 |
| 1147 | 126 | A | G | C | A | T | A | 250 | 111/113 |
| 1280 | 116/126 | A | G | C | A | T | A | 250/254 | 111 |
| 1299 | 116/126 | A | G | C | A | T | A | 250 | 111 |
| 1340 | 116/126 | A | G | C | A | T | … | 250 | 111/113 |
| 1479 | 116 | A | G | C | A | T | A | 250 | 111 |
| 2456 | 116/126 | A | G | C | A | T | A | 250 | 111 |
| 2463 | 126 | A | G | C | A | T | A | 250 | … |
| 2499 | 126 | A | G | C | A | T | A | 250 | 115 |
| 2502 | 118 | A | G | C | A | T | A | 250 | 111/113 |
| 2505 | 126 | A | G | C | A | T | A | 250 | 115 |
| 2517 | 116/118 | A | G | C | A | T | A | 250 | … |
| 2572 | 126 | A | G | C | A | T | A | 250 | 107 |
| 2579 | 126 | A | G | C | A | T | A | 250 | 111 |
| 2580 | 126 | A | G | C | A | T | A | 250 | 111 |
| 2584 | 126 | A | G | C | A | T | A | 250 | 113/115 |
| 2725 | 126 | A | G | C | A | T | A | 250/252 | 113 |
| 2728 | 126 | A | G | C | A | T | A | 250 | 111 |
| 1155 | 116/126 | A | G/A | C | A | T | A | 250 | 111/115 |
| 2455 | 126 | A | G/A | C | A | T | A | 250 | 111/113 |
| 1870 | 116/126 | A | G | C | A/G | T | A | 250/252 | 107/111 |
| 2041 | 118 | A | G | … | A/G | T | A/G | 250 | … |
| 2561 | 116/126 | A | G/A | C/T | A/G | T/G | A/G | 250 | 111/113 |
| 2573 | 126 | A | G | C/T | A/G | T/G | A/G | 250 | 111 |
| 2625 | 116 | A/G | G/A | C | A/G | T | A/G | 250/256 | 111 |
| Denmark: | |||||||||
| D2 | 116 | A | G | C | A | T | A | 250 | 113 |
| 116 | A | G | C | A | T | A | 250 | 113 | |
| D4 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| D13 | 116 | A | G | C | A | T | A | 250 | 113 |
| 116 | A | G | C | A | T | A | 250 | 113 | |
| D18 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 113 | |
| D19 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| D40 | 116 | A | G | C | A | T | A | 250 | 111/113 |
| 126 | A | G | C | A | T | A | 250 | ||
| D44 | 116 | A | G | C | A | T | A | 250 | 111 |
| 116 | A | G | C | A | T | A | 250 | 113 | |
| D5 | 126 | A | G | C | A | T | A | 250 | 113 |
| D7 | 126 | A | G | C | A | T | A | 250 | 111 |
| D8 | 116/126 | A | G | C | A | T | A | 250 | 107/113 |
| D10 | 126 | A | G | C | A | T | A | 250 | 111/113 |
| D15 | 116 | A | G | C | A | T | A | 250 | 113 |
| D16 | 116/126 | A | G | C | A | T | A | 250 | 111 |
| D22 | 116/126 | A | G | C | A | T | A | 250 | 111 |
| D24 | 116 | A | G | C | A | T | A | 250 | 111/113 |
| D39 | 116/118 | A | G | C | A | T | A | 250 | 111/115 |
| D32 | 116/126 | A | G | C | A/G | T | A/G | … | 111/115 |
| D11 | 116/126 | A/G | G/A | C/T | A | T | A | 250 | 111/115 |
| D17 | 118/126 | A | G/A | C/T | A/G | T | A | 250 | 107/111 |
| D25 | 116 | A | G | C/T | A/G | T | A/G | 250 | 109/113 |
| D31 | 116 | A | G | C | A/G | T/G | A/G | 250 | 113/115 |
| D23 | 116/126 | A | G | C/T | A/G | T/G | A/G | 250 | 111 |
| The Netherlands: | |||||||||
| 1003 | 126 | A | G | C | A | T | A | 250 | 115 |
| 126 | A | G | C | A | T | A | 250 | 115 | |
| 1011 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| 1370 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 113 | |
| 1374 | 126 | A | G | C | A | T | A | 250 | 115 |
| 126 | A | G | C | A | T | A | 250 | 115 | |
| 1404 | 126 | A | G | C | A | T | A | 250 | 113 |
| 126 | A | G | C | A | T | A | 250 | 115 | |
| 1298 | 126 | A | G | C | A | T | A | 250 | 115 |
| 1375 | 126 | A | G | C | A | T | A | 250 | 111 |
| 1386 | 128 | A | G | C | A | T | A | 250 | 111 |
| 1387 | 126 | A | G | C | A | T | A | 250 | 113 |
| 1415 | 126 | A | G | C | A | T | A | 250 | 115 |
| 1044 | 116 | A | G | C | A | T | A | 250 | 111 |
| 1238 | 126 | A | G | C | A | T | A | 250 | 113 |
| 1420 | 118/126 | A | G | C | A | T | A | 250 | 111 |
| 776 | 126 | A | G | C | A | T | A | 250 | 107/113 |
| 1008 | 126 | A | G | C | A | T | A | 250 | 111/115 |
| 1368 | 126 | A | G | C | A | T | A | 250 | 111/113 |
| 956 | 126 | A/G | G/A | C | A | T | A/G | 250 | 111 |
| United Kingdom:b | |||||||||
| UK1 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| UK2 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| UK5 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| UK7 | 116/126 | A | G | C | A | T | A | 250 | 111/113 |
| UK3 | 116/126 | A | G | C | A/G | … | A/G | … | 107/111 |
| UK4 | 116/126 | A | G | C | A | … | A | … | 113 |
| UK6 | 126 | A/G | G/A | C | A | T | A | … | 111 |
| UK8 | 116/126 | A/G | G/A | C/T | A/G | T/G | A/G | … | 107/111 |
| UK10 | 116/126 | A | G/A | C | A | T | A | … | 111/115 |
| UK11c | 116/126 | A/G | G/A | … | A/G | … | … | … | … |
| UK12 | 116/126 | A | G/A | C | A | T | A | … | … |
| Spain: | |||||||||
| SP1 | 126 | A | G | C | A | T | A | 250 | 113 |
| 126 | A | G | C | A | T | A | 250 | 113 | |
| SP2 | 116 | A | G | C | A | T | A | 250 | 111 |
| 116 | A | G | C | A | T | A | 250 | 111 | |
| SP3 | 116 | A | G | C | A | T | A | 250 | 111 |
| 116 | A | G | C | A | T | A | 250 | 115 | |
| SP5 | 126 | A | G | C | A | T | A | 250 | 111/115 |
| 1076 | 126 | A | G | C | A | T | A | 250 | 111 |
| SP4 | 116/118 | A/G | G/A | C/T | A/G | T/G | A/G | 246/252 | 113 |
| Sweden: | |||||||||
| S1 | 116 | A | G | C | A | T | A | 250 | 113 |
| 126 | A | G | C | A | T | A | 250 | 113 | |
| S2 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| S3 | 116 | A | G | C | A | T | A | 250 | 113 |
| 126 | A | G | C | A | T | A | 250 | 113 | |
| S4 | 116 | A | G/A | C | A | T | A | 250 | 111/115 |
| S5 | 116 | A/G | G/A | C/T | A | T | A | 250 | 111/113 |
| S6 | 116/126 | A/G | G/A | C/T | A | T | A | 250 | 113 |
| Norway: | |||||||||
| N4 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| N5 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| N25 | 116/126 | A | G | C | A | T | A | 250 | 111/113 |
| N15 | 116/126 | A | G | C | A/G | T/G | A/G | 250 | 109/111 |
| Belgium: | |||||||||
| 1862 | 116 | A | G | C | A | T | A | 250 | 111 |
| 116 | A | G | C | A | T | A | 250 | 111 | |
| 1863 | 126 | A | G | C | A/G | T/G | A/G | 250 | 115 |
| Canada: | |||||||||
| 1158 | 128 | A | G/A | C | A | T | A | 250 | 111 |
| Colombia: | |||||||||
| 1150 | 116 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
| France: | |||||||||
| 1223 | 116 | A | G | C | A | T | A | 250 | 111 |
| 116 | A | G | C | A | T | A | 250 | 113 | |
| Germany: | |||||||||
| 1294 | 116/118 | A/G | G/A | C/T | A/G | T/G | A/G | 250 | … |
| South Africa: | |||||||||
| 1128 | 126 | A | G | C | A | T | A | 250 | 111 |
| 126 | A | G | C | A | T | A | 250 | 111 | |
Note.— Genotyping was incomplete in some patients because of a lack of DNA material.
Patient was African American.
Samples of genomic DNA of the patients in the United Kingdom were provided by Dr. Xue Zhong Liu.
Patient was Chinese.
In all the 2299delG alleles for which the phase could be determined (116 of 148), the 2299delG mutation was associated with one core haplotype, A-G-C-A-T-A. In the remaining 32 alleles, other haplotypes for the 2299delG allele would be formally possible; however, the A-G-C-A-T-A core haplotype could not be excluded. Except in one Spanish patient (SP4), the core haplotype could be extended on the 3′ side, to span 325 kb (A-G-C-A-T-A-250), including the marker AFM268ZD1.
Twelve different USH2A SNP haplotypes were observed in a panel of normal chromosomes from Norwegian nuclear families (n=108); four major haplotypes (1–4) had frequencies of .60, .13, .06, and .03, and eight derivative haplotypes showed frequencies <.03. These haplotype frequencies were not significantly different from those observed in chromosomes from parents of patients with Usher syndrome (n=49) (table 2).
Table 2.
Frequencies of 2299delG and Normal Haplotypes
|
Genotype ata |
Frequencies of |
||||||||
| Haplotype | 373A/G(.76/.24) | 504G/A(.72/.28) | 1419C/T(.78/.22) | IVS15+35A/G(.66/.34) | IVS17−8T/G(.76/.24) | 4457A/G(.64/.36) | NormalControlAlleles(n = 108) | NormalParentalAlleles(n = 49) | 2299delGAlleles(n= 116) |
| 1 | A | G | C | A | T | A | .60 | .53 | 1.00b |
| 2 | G | A | T | G | G | G | .13c | .22c | |
| 3 | A | G | C | G | G | G | .06 | .02 | |
| 4 | G | A | C | A | T | A | .05 | .04 | |
| 5–12d | … | … | … | … | … | … | .16 | .19 | |
Genotypes are given according to SNPs (frequencies).
Statistical significance of the association was measured by the χ2 test ( χ2 = 57.2, P < .001).
The frequency of haplotype 2 is higher than expected on the basis of the individual allele frequencies and illustrates a situation of strong linkage disequilibrium, (χ2 = 42.0, P < .001).
Eight haplotypes with frequencies <.03.
Because of the wide geographic distribution of the mutation and because of the haplotype heterogeneity at the microsatellite markers (AFM144XF2, AFM268ZD1, and AFM143XF10) that flank the USH2A locus, it had been previously suggested that the 2299delG mutation did not arise in a common ancestor (Eudy et al. 1998; Liu et al. 1999). In our analysis, however, all 116 2299delG chromosomes for which the phase was known were found to be in complete association with one specific core haplotype, which was designated “USH2A haplotype 1.” Statistical significance of the association was demonstrated using the χ2 test (χ2=57.2; P<.001). The data strongly indicate that the 2299delG alleles originated from an ancestral mutational event. The variation at the flanking markers, especially the AFM144XF2 and AFM143XF10, among the 2299delG chromosomes probably reflects the generally high mutation rate (10−3 to 10−4 per locus per generation) at microsatellite sites (Weber and Wong 1993).
The fact that the 2299delG-associated haplotype is also the most frequent in the unaffected population suggests that the genetic distance between these two groups is low. All but two of the patients in this study are European or of European descent. It is tempting to speculate that the 2299delG mutation results from an old mutational event that happened to arise on the most common haplotype in the present European genetic background and that was spread, through migration and subsequent founder effects, throughout Europe. Furthermore, the 2299delG mutation was probably brought to the New World in recent times, because it occurs in countries with a history of European immigration. This definitely applies to the United States and to the singleton patients in South Africa and Canada. However, two notable exceptions are the American patient of African ancestry (patient 791) and the patient of Chinese ancestry (UK11, for whom the phase could not be determined). Both patients are heterozygous for the 2299delG mutation. A DNA sample from a Chinese patient who has been reported to be homozygous for the 2299delG mutation (Liu et al. 1999) was not available for this study. Because no one has reported studies of 2299delG mutation frequencies in non-European populations, it remains to be elucidated whether 2299delG is present in these two patients as a result of recent ethnic admixture or is present in the gene pool of other populations such as those of Africa and China.
In summary, our data provide the first molecular evidence that the frequent and widespread 2299delG mutation results from an ancient mutation of common origin rather than from multiple recurrent mutational events on a common haplotype. Although all the 2299delG alleles studied contain a conserved SNP core haplotype that spans >250 kb, the haplotype varies widely, and the association between the 2299delG mutation and the haplotypes of the flanking microsatellite markers appears to be random. Regarding the physical distances between these markers, this variability cannot be explained by recombination alone (fig. 1). Thus, the above data clearly demonstrate that, because of their relatively high mutation frequency, microsatellites must be applied in combination with SNP markers when studying the origin and distribution of mutation.
Acknowledgments
This work has been supported by Norwegian Foundation for Health and Rehabilitation grant 1998/257 (B.D., Ø.N., and L.T.) and a grant from the Danish Support Foundation for the Blind (T.R.). The study was approved by the Regional Research Ethics Committee in Tromsø. We would like to thank Dr. Xue Zhong Liu, for providing DNA samples from UK/China, and Dr. Carmen Ayuso, for providing Spanish DNA control samples.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Genome Database, http://www.gdb.org (for genotyping of markers)
- Hereditary Hearing Loss Home Page (G. Van Camp, R. J. H. Smith), http://www.uia.ac.be/dnalab/hhh/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for Usher syndrome [MIM 276900])
References
- Adato A, Weston MD, Berry A, Kimberling WJ, Bonne-Tamir A (2000) Three novel mutations and twelve polymorphisms identified in the USH2A gene in Israeli USH2 families. Hum Mutat 15:388 [DOI] [PubMed] [Google Scholar]
- Beneyto MM, Cuevas JM, Millan JM, Espinos C, Mateu E, Gonzalez-Cabo P, Baiget M, Domenech M, Bernal S, Ayuso C, Garcia-Sandoval B, Trujillo MJ, Borrego S, Antinolo G, Carballo M, Najera C (2000) Prevalence of 2314delG mutation in Spanish patients with Usher syndrome type II (USH2). Ophthalmic Genet 21:123–128 [PubMed] [Google Scholar]
- Boughman JA, Vernon M, Shaver KA (1983) Usher syndrome: definition and estimate of prevalence from two high-risk populations. J Chronic Dis 36:595–603 [DOI] [PubMed] [Google Scholar]
- Dreyer B, Tranebjærg L, Rosenberg T, Weston MD, Kimberling WJ, Nilssen Ø (2000) Identification of novel USH2A mutations: implications for the structure of USH2A protein. Eur J Hum Genet 8:500–506 [DOI] [PubMed] [Google Scholar]
- Eudy JD, Weston MD, Yao S, Hoover DM, Rehm HL, Ma-Edmonds M, Yan D, Ahmad I, Cheng JJ, Ayuso C, Cremers C, Davenport S, Möller C, Talmadge CB, Beisel KW, Tamayo M, Morton CC, Swaroop A, Kimberling WJ, Sumegi J (1998) Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science 280:1753–1757 [DOI] [PubMed] [Google Scholar]
- Liu X-Z, Hope C, Liang CY, Zou JM, Xu LR, Cole T, Mueller RF, Bundey S, Nance W, Steel KP, Brown SDM (1999) A mutation (2314delG) in the Usher syndrome type IIA gene: high prevalence and phenotypic variation. Am J Hum Genet 64:1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C, Sweklo EA, Berson EL, Dryja TP (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 66:1975–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg T, Haim M, Hauch AM, Parving A (1997) The prevalence of Usher syndrome and other retinal dystrophy–hearing impairment associations. Clin Genet 51:314–321 [DOI] [PubMed] [Google Scholar]
- Smith RJ, Berlin CI, Hejtmancik JF, Keats BJ, Kimberling WJ, Lewis RA, Möller CG, Pelias MZ, Tranebjærg L (1994) Clinical diagnosis of the Usher syndromes: Usher Syndrome Consortium. Am J Med Genet 50:32–38 [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128 [DOI] [PubMed] [Google Scholar]
- Weston MD, Eudy JD, Fujita S, Yao S, Usami S, Cremers C, Greenburg J, Ramesar R, Martini A, Möller C, Smith RJ, Sumegi J, Kimberling WJ (2000) Genomic structure and identification of novel mutations in Usherin, the gene responsible for Usher syndrome type IIa. Am J Hum Genet 66:1199–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]